Climatology variable A
Jens Daniel Müller
24 October, 2022
Last updated: 2022-10-24
Checks: 7 0
Knit directory: emlr_mod_preprocessing/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200707) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e855cad. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: code/backup_analysis_scripts/Cant_CB_annual_all_models_backup_20220704.Rmd
Unstaged changes:
Modified: code/Workflowr_project_managment.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/Climatology_A_2007_all_models.Rmd) and HTML
(docs/Climatology_A_2007_all_models.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | e829a11 | jens-daniel-mueller | 2022-07-05 | Build site. |
| Rmd | 923e07e | jens-daniel-mueller | 2022-07-04 | rerun for all models |
| html | 89cbb74 | jens-daniel-mueller | 2022-05-26 | Build site. |
| Rmd | 379a012 | jens-daniel-mueller | 2022-05-26 | rerun for all models |
| html | 9e85627 | jens-daniel-mueller | 2022-05-24 | Build site. |
| Rmd | 30f99f7 | jens-daniel-mueller | 2022-05-24 | rerun only for planktom12 with factor /100 for po4 |
| html | 42bbb89 | jens-daniel-mueller | 2022-05-24 | Build site. |
| Rmd | e82a302 | jens-daniel-mueller | 2022-05-23 | bug fixes for 5 models |
| html | 40863a2 | jens-daniel-mueller | 2022-05-22 | Build site. |
| Rmd | d9d4d68 | jens-daniel-mueller | 2022-05-21 | rebuild with revised output structure and all models |
| html | 2257c34 | jens-daniel-mueller | 2022-05-20 | Build site. |
| html | 31ae80a | jens-daniel-mueller | 2022-05-20 | Build site. |
| Rmd | b6d47dd | jens-daniel-mueller | 2022-05-20 | write files without 3D_ALL insert |
| html | 0dd6813 | jens-daniel-mueller | 2022-05-15 | Build site. |
| Rmd | 1e98fee | jens-daniel-mueller | 2022-05-15 | included more models excp. planktom12 |
| html | 06054b2 | jens-daniel-mueller | 2022-05-10 | Build site. |
| Rmd | d43e8c0 | jens-daniel-mueller | 2022-05-10 | rerun all with multi model subsetting |
1 Select basin mask
# use only three basin to assign general basin mask
# ie this is not specific to the MLR fitting
basinmask <- basinmask %>%
filter(MLR_basins == "2") %>%
select(lat, lon, basin_AIP)2 Read nc file
Here we used annual output of cmorized (1x1) model with variable forcing (RECCAP2 RunA) in year 2007 as the predictor climatology. Predictors include:
- Salinity (sal)
- Potential temperature (theta, not predictor, used for temperature calculation)
- In-situ temperature (temp, calculated)
- DIC (tco2)
- ALK (talk)
- oxygen
- AOU (calculated)
- nitrate
- phosphate
- silicate
# set name of model to be subsetted
model_ID <- "A"
# for loop across variables
variables <-
c("so", "thetao", "dissic", "talk", "o2", "no3", "po4", "si")
models <- list.files(path_cmorized)
models <-
models[!str_detect(models, pattern = "\\.t|\\.z")]
models <-
models[str_detect(
models,
pattern = c(
"CESM|CNRM|EC-Earth3|FESOM_REcoM_LR|MOM6-Princeton|MRI-ESM2-1|NorESM-OC1.2|ORCA025-GEOMAR|ORCA1-LIM3-PISCES|planktom12"
)
)]
# depth levels not available
models <-
models[!str_detect(models, pattern = "CNRM")]
# files chunked into decades
models <-
models[!str_detect(models, pattern = "ORCA025-GEOMAR")]
# "so" variable only NA's
models <-
models[!str_detect(models, pattern = "MOM6-Princeton")]# load python scripts
source_python(paste0(
path_functions,
"python_scripts/Gamma_GLODAP_python.py"
))# models <- models[7]
for (i_model in models) {
# i_model <- models[1]
print(i_model)
variables_available <-
list.files(path = paste0(path_cmorized, i_model),
pattern = paste0("_", model_ID, "_"))
variables_available <-
str_split(variables_available,
pattern = "_",
simplify = TRUE)[, 1]
variables_available <-
variables_available[variables_available %in% variables]
variables_available <- unique(variables_available)
print(variables_available)
for (i_variable in variables_available) {
# i_variable <- variables_available[3]
# read list of all files
file <-
list.files(path = paste0(path_cmorized, i_model),
pattern = paste0(i_variable, "_"))
file <-
file[str_detect(file, pattern = paste0("_", model_ID, "_"))]
print(file)
# read in data
if (i_model == "NorESM-OC1.2_3D_ALL_v20211125") {
variable_data <-
tidync::tidync(paste(paste0(path_cmorized, i_model),
file,
sep = "/"))
} else {
variable_data <-
read_ncdf(paste(paste0(path_cmorized, i_model),
file,
sep = "/"),
make_units = FALSE)
}
if (i_model == "CESM-ETHZ_3D_ALL_v20211122") {
variable_data <-
variable_data %>% slice(index = 28, along = "time_ann")
} else if (i_model == "FESOM_REcoM_LR_3D_all_v20211119") {
variable_data <-
variable_data %>% slice(index = 28, along = "Time")
} else if (i_model == "planktom12_3d_all_v20220404") {
variable_data <-
variable_data %>% slice(index = 28, along = "TIME")
} else if (i_model == "MRI-ESM2-1_3D_ALL_v20220502" &
i_variable == "si") {
variable_data <-
variable_data %>% slice(index = 1, along = "time")
} else if (i_model == "NorESM-OC1.2_3D_ALL_v20211125") {
variable_data <- variable_data %>%
tidync::hyper_filter(time = index == 28) %>%
tidync::hyper_tibble() %>%
select(-time)
} else {
variable_data <- variable_data %>% slice(index = 28, along = "time")
}
print(variable_data)
# convert to tibble
variable_data_tibble <- variable_data %>%
as_tibble()
# remove open link to nc file
rm(variable_data)
# remove na values
variable_data_tibble <-
variable_data_tibble %>%
filter(!is.na(!!sym(i_variable)))
if (i_model == "CNRM-ESM2-1_3D_ALL_v20211208") {
variable_data_tibble <- variable_data_tibble %>%
rename(depth = lev)
}
if (i_model == "FESOM_REcoM_LR_3D_all_v20211119") {
variable_data_tibble <- variable_data_tibble %>%
rename(
lat = Lat,
lon = Lon,
depth = Depth
)
}
if (i_model == "MOM6-Princeton_3D_ALL_v20220125") {
variable_data_tibble <- variable_data_tibble %>%
rename(depth = z_l)
}
if (i_model == "MRI-ESM2-1_3D_ALL_v20220502") {
variable_data_tibble <- variable_data_tibble %>%
rename(depth = lev)
}
if (i_model == "planktom12_3d_all_v20220404") {
variable_data_tibble <- variable_data_tibble %>%
rename(lon = LONGITUDE,
lat = LATITUDE,
depth = DEPTH)
}
if (i_model == "planktom12_3d_all_v20220404" &
i_variable == "po4") {
variable_data_tibble <-
variable_data_tibble %>%
mutate(po4 = po4 / 100)
}
# harmonize longitudes
variable_data_tibble <- variable_data_tibble %>%
mutate(lon = if_else(lon < 20, lon + 360, lon))
# only consider model grids within basinmask
variable_data_tibble <-
inner_join(variable_data_tibble, basinmask) %>%
select(-basin_AIP)
if (exists("climatology")) {
climatology <- full_join(climatology, variable_data_tibble)
}
if (!exists("climatology")) {
climatology <- variable_data_tibble
}
}
if (i_model == "FESOM_REcoM_LR_3D_all_v20211119") {
climatology <- climatology %>%
mutate(po4 = no3/16)
}
if (i_model == "planktom12_3d_all_v20220404") {
climatology <- climatology %>%
mutate(no3 = po4*16)
}
climatology <- climatology %>%
rename(
sal = so,
THETA = thetao,
tco2 = dissic,
talk = talk,
oxygen = o2,
nitrate = no3,
phosphate = po4,
silicate = si
)
climatology <- climatology %>%
mutate(depth = round(depth))
# In-situ temperature
climatology <- climatology %>%
mutate(temp = gsw_pt_from_t(
SA = sal,
t = THETA,
p = 10.1325,
p_ref = depth
))
# Example profile from North Atlantic Ocean
print(
climatology %>%
filter(lat == params_global$lat_Atl_profile,
lon == params_global$lon_Atl_section) %>%
ggplot() +
geom_line(aes(temp, depth, col = "insitu")) +
geom_point(aes(temp, depth, col = "insitu")) +
geom_line(aes(THETA, depth, col = "theta")) +
geom_point(aes(THETA, depth, col = "theta")) +
scale_y_reverse() +
labs(title = i_model) *
scale_color_brewer(palette = "Dark2", name = "Scale"))
# Unit conversion
# Model results are given in [mol m^-3^], whereas GLODAP data are in [µmol kg^-1^].
# For comparison, model results were converted from [mol m^-3^] to [µmol kg^-1^]
climatology <- climatology %>%
mutate(
rho = gsw_pot_rho_t_exact(
SA = sal,
t = temp,
p = depth,
p_ref = 10.1325
),
tco2 = tco2 * (1e+06 / rho),
talk = talk * (1e+06 / rho),
oxygen = oxygen * (1e+06 / rho),
nitrate = nitrate * (1e+06 / rho),
phosphate = phosphate * (1e+06 / rho),
silicate = silicate * (1e+06 / rho)
)
# AOU calculation
climatology <- climatology %>%
mutate(
oxygen_sat_m3 = gas_satconc(
S = sal,
t = temp,
P = 1.013253,
species = "O2"
),
oxygen_sat_kg = oxygen_sat_m3 * (1e+3 / rho),
AOU = oxygen_sat_kg - oxygen
) %>%
select(-oxygen_sat_kg, -oxygen_sat_m3)
# Neutral density calculation
# Neutral density gamma was calculated with a Python script provided by Serazin et al (2011)
# which performs a polynomial approximation of the original gamma calculation.
#calculate pressure from depth
climatology <- climatology %>%
mutate(CTDPRS = gsw_p_from_z(-depth,
lat))
# rename variables according to python script
climatology_gamma_prep <- climatology %>%
rename(LATITUDE = lat,
LONGITUDE = lon,
SALNTY = sal)
# calculate gamma
climatology_gamma_calc <-
calculate_gamma(climatology_gamma_prep)
# reverse variable naming
climatology <- climatology_gamma_calc %>%
select(-c(CTDPRS, THETA)) %>%
rename(
lat = LATITUDE,
lon = LONGITUDE,
sal = SALNTY,
gamma = GAMMA
)
climatology <- as_tibble(climatology)
rm(climatology_gamma_calc, climatology_gamma_prep)
# write_climatology_predictor_file
# select relevant columns
climatology <- climatology %>%
select(-c(rho))
# Remove 3D_all from model name
i_model <- str_remove(i_model, "3D_ALL_|3D_all_|3d_all_")
# write csv file
climatology %>%
write_csv(paste0(path_preprocessing,
i_model,
"_climatology_runA_2007.csv"))
rm(climatology)
}[1] "CESM-ETHZ_3D_ALL_v20211122"
[1] "dissic" "no3" "o2" "po4" "si" "so" "talk" "thetao"
[1] "dissic_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
dissic 1.53844 2.063345 2.121567 2.112429 2.172408 2.338243 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375
[1] "no3_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
no3 0 0.0006632728 0.003444489 0.007649824 0.0142726 0.02791386 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375
[1] "o2_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
o2 0.1949853 0.2097159 0.2504627 0.2672393 0.3263081 0.3876033 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375
[1] "po4_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
po4 0 9.326071e-05 0.0002925115 0.0005661223 0.0009729294 0.001930219 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375
[1] "si_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
si 0.0007265678 0.003127281 0.005360665 0.01249062 0.01393934 0.1167264 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375
[1] "so_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
so 17.77432 34.11263 34.72358 34.70072 35.68783 40.53304 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375
[1] "talk_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
talk 1.754645 2.341156 2.360543 2.36032 2.404855 2.692481 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375
[1] "thetao_CESM-ETHZ_A_1_gr_1980-2018_v20211122.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
thetao -1.782697 2.924084 16.20286 14.43304 25.42252 30.07337 34720
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 60 NA NA NA NA [60] 5,...,5375 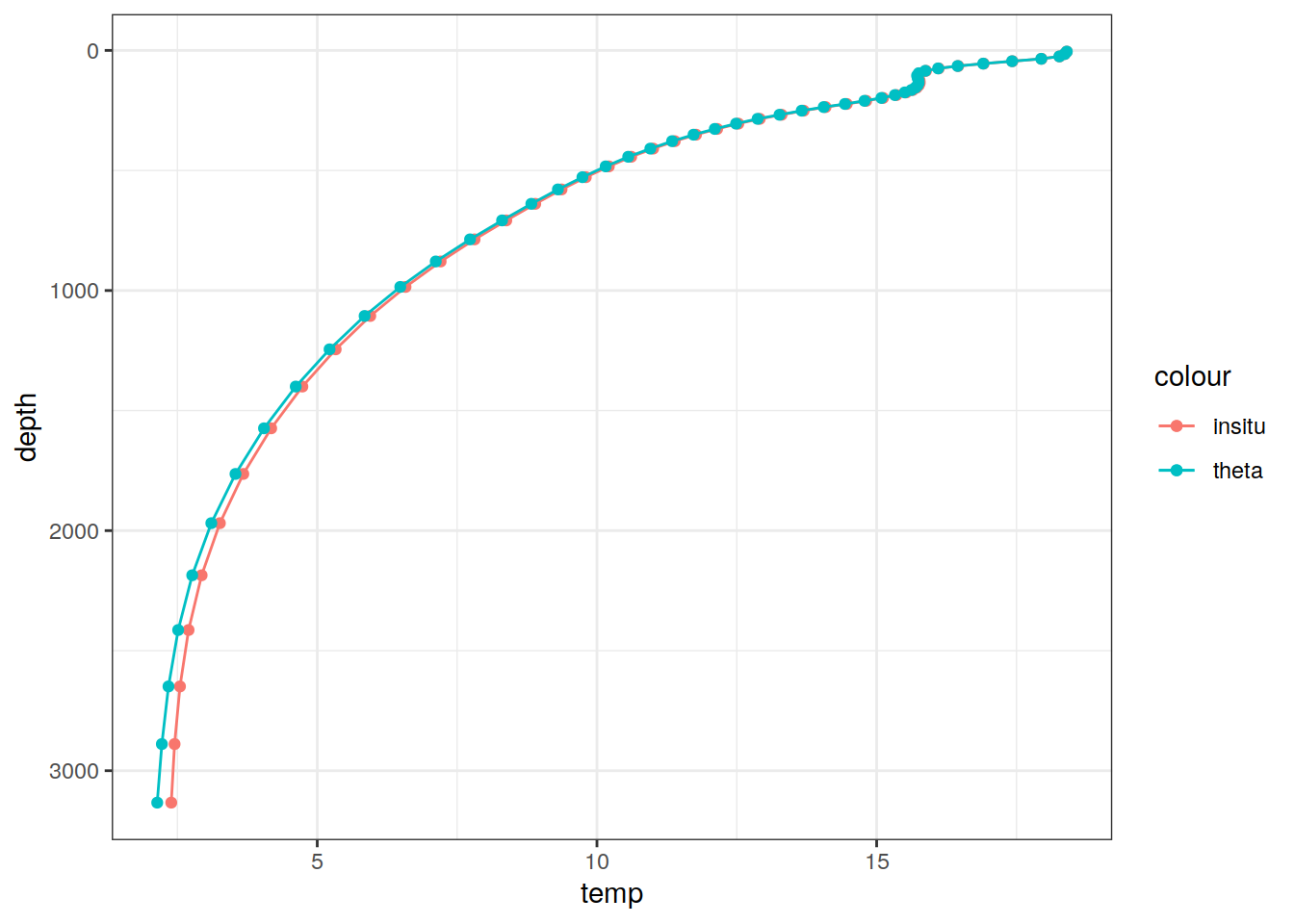
[1] "EC-Earth3_3D_ALL_v20220323"
[1] "dissic" "no3" "o2" "po4" "si" "so" "talk" "thetao"
[1] "dissic_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
dissic 0.5402265 2.007797 2.065164 2.071406 2.13433 2.557395 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "no3_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
no3 5.910887e-07 0.0002337237 0.004086725 0.008991736 0.01686225 0.03196622
NA's
no3 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "o2_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
o2 0.1961801 0.2094422 0.2514559 0.2703499 0.331589 0.4155927 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "po4_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu.
po4 2.666838e-07 0.0001178296 0.0004282285 0.0007320858 0.001306314
Max. NA's
po4 0.004568885 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "si_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
si 0.0007297698 0.001440996 0.002759601 0.01274259 0.01048773 0.1026836 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "so_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
so 4.976272 33.89646 34.33475 34.29951 35.23156 40.07595 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "talk_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
talk 0.5222299 2.282914 2.313059 2.304122 2.338625 2.834188 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "thetao_EC-Earth3_A_1_gr_1980-2018_v20220323.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
thetao -1.90141 2.354912 15.96602 14.36113 25.54307 30.26387 34235
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229) 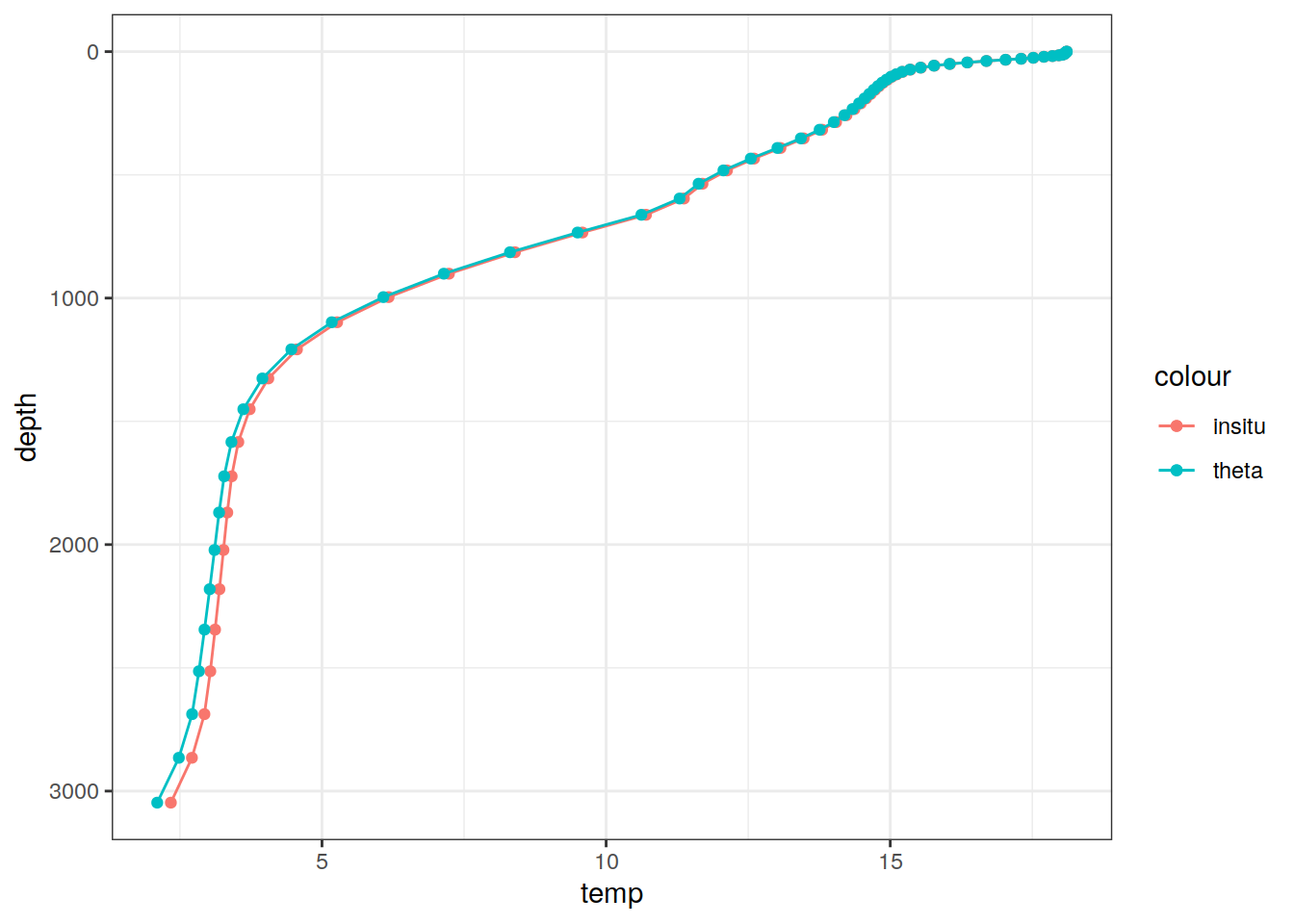
[1] "FESOM_REcoM_LR_3D_all_v20211119"
[1] "dissic" "no3" "o2" "si" "so" "talk" "thetao"
[1] "dissic_FESOM_REcoM_LR_A_1_gr_1980-2018_v20211119.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
dissic 1.397473 1.980928 2.056114 2.062378 2.139095 2.243466 33173
dimension(s):
from to offset delta refsys point values x/y
Lon 1 360 0 1 NA NA NULL [x]
Lat 1 180 -90 1 NA NA NULL [y]
Depth 1 46 NA NA NA NA [46] 0,...,5900
[1] "no3_FESOM_REcoM_LR_A_1_gr_1980-2018_v20211119.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
no3 1.154229e-05 3.847795e-05 0.002167632 0.007301974 0.01327029 0.04178772
NA's
no3 33173
dimension(s):
from to offset delta refsys point values x/y
Lon 1 360 0 1 NA NA NULL [x]
Lat 1 180 -90 1 NA NA NULL [y]
Depth 1 46 NA NA NA NA [46] 0,...,5900
[1] "o2_FESOM_REcoM_LR_A_1_gr_1980-2018_v20211119.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
o2 0.1862063 0.2125652 0.2526134 0.2630829 0.3142638 0.367201 33173
dimension(s):
from to offset delta refsys point values x/y
Lon 1 360 0 1 NA NA NULL [x]
Lat 1 180 -90 1 NA NA NULL [y]
Depth 1 46 NA NA NA NA [46] 0,...,5900
[1] "si_FESOM_REcoM_LR_A_1_gr_1980-2018_v20211119.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
si 1.656723e-06 2.832228e-05 9.510469e-05 0.00372529 0.0008885778 0.06004048
NA's
si 33173
dimension(s):
from to offset delta refsys point values x/y
Lon 1 360 0 1 NA NA NULL [x]
Lat 1 180 -90 1 NA NA NULL [y]
Depth 1 46 NA NA NA NA [46] 0,...,5900
[1] "so_FESOM_REcoM_LR_A_1_gr_1980-2018_v20211119.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
so 18.7434 33.80206 34.3102 34.31283 35.22783 39.38908 33173
dimension(s):
from to offset delta refsys point values x/y
Lon 1 360 0 1 NA NA NULL [x]
Lat 1 180 -90 1 NA NA NULL [y]
Depth 1 46 NA NA NA NA [46] 0,...,5900
[1] "talk_FESOM_REcoM_LR_A_1_gr_1980-2018_v20211119.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
talk 1.336164 2.276783 2.30378 2.29198 2.317252 2.571417 33173
dimension(s):
from to offset delta refsys point values x/y
Lon 1 360 0 1 NA NA NULL [x]
Lat 1 180 -90 1 NA NA NULL [y]
Depth 1 46 NA NA NA NA [46] 0,...,5900
[1] "thetao_FESOM_REcoM_LR_A_1_gr_1980-2018_v20211119.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
thetao -1.891329 2.555371 15.80864 14.07006 24.94776 29.80794 33173
dimension(s):
from to offset delta refsys point values x/y
Lon 1 360 0 1 NA NA NULL [x]
Lat 1 180 -90 1 NA NA NULL [y]
Depth 1 46 NA NA NA NA [46] 0,...,5900 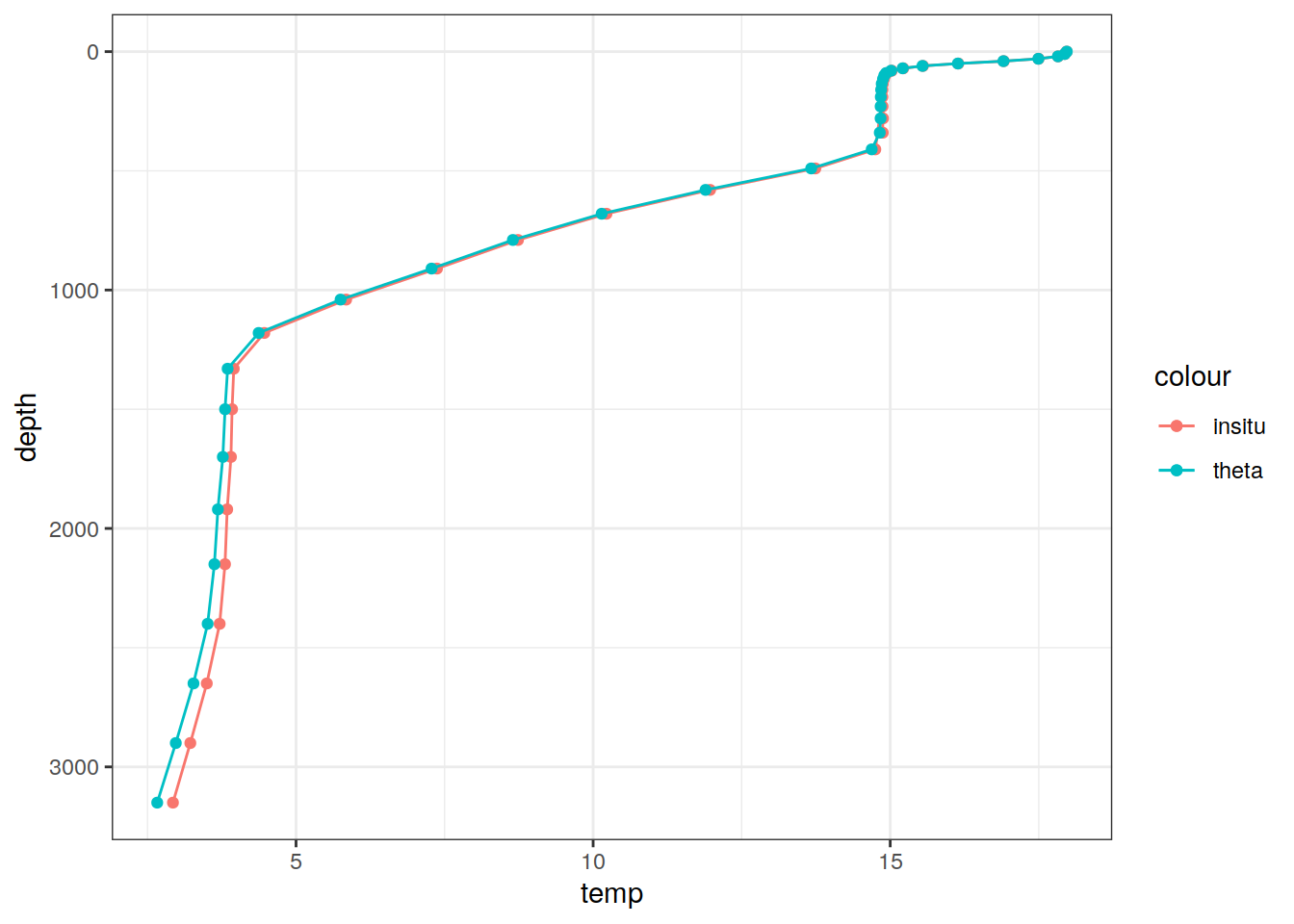
[1] "MRI-ESM2-1_3D_ALL_v20220502"
[1] "dissic" "no3" "o2" "po4" "si" "so" "talk" "thetao"
[1] "dissic_MRI-ESM2-1_A_1_gr_1980-2018_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
dissic 0.4204865 1.969964 2.043 2.048039 2.140602 2.266817 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550)
[1] "no3_MRI-ESM2-1_A_1_gr_1980-2018_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
no3 4.240297e-06 0.0001957148 0.00809673 0.0105852 0.02124245 0.04277238 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550)
[1] "o2_MRI-ESM2-1_A_1_gr_1980-2018_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
o2 0.1543084 0.2109115 0.2528302 0.2677437 0.324883 0.3961899 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550)
[1] "po4_MRI-ESM2-1_A_1_gr_1980-2018_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
po4 9.871876e-05 0.0002635115 0.000744225 0.0009022392 0.001568535 0.002918553
NA's
po4 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550)
[1] "si_MRI-ESM2-1_A_1_gr_1980-1980_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
si 0.0004158035 0.00200341 0.003626204 0.01160255 0.009417075 0.09425249 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550)
[1] "so_MRI-ESM2-1_A_1_gr_1980-2018_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
so 6.063022 34.13029 34.58293 34.49567 35.52903 40.81694 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550)
[1] "talk_MRI-ESM2-1_A_1_gr_1980-2018_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
talk 0.403091 2.257826 2.322342 2.285143 2.347492 2.656055 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550)
[1] "thetao_MRI-ESM2-1_A_1_gr_1980-2018_v20220502.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
thetao -1.745155 2.44172 15.76996 14.27788 25.47815 30.01773 31002
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
lev 1 61 NA NA NA FALSE [0,2),...,[6500,6550) 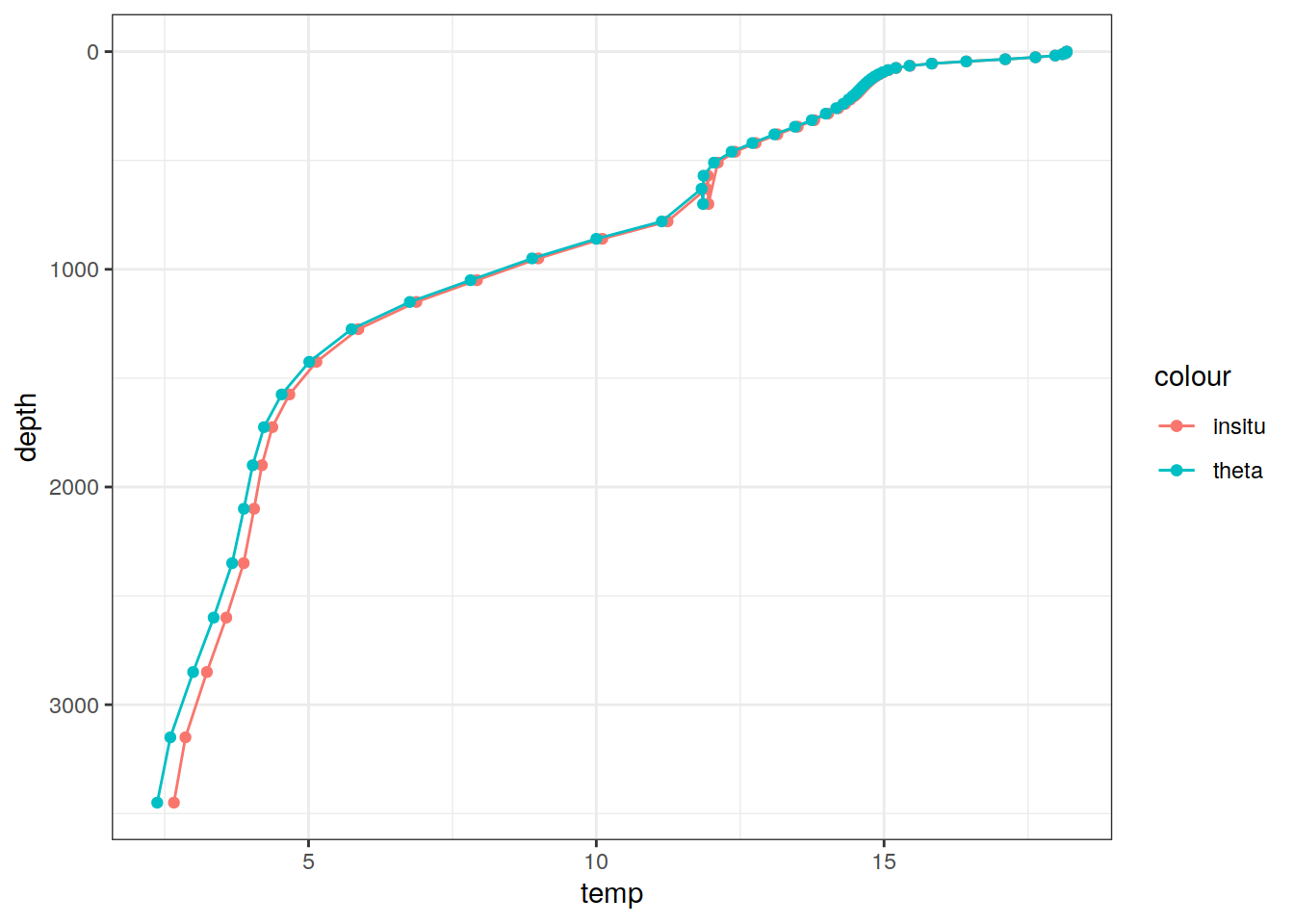
| Version | Author | Date |
|---|---|---|
| e829a11 | jens-daniel-mueller | 2022-07-05 |
| 89cbb74 | jens-daniel-mueller | 2022-05-26 |
| 9e85627 | jens-daniel-mueller | 2022-05-24 |
| 42bbb89 | jens-daniel-mueller | 2022-05-24 |
| 40863a2 | jens-daniel-mueller | 2022-05-22 |
| 31ae80a | jens-daniel-mueller | 2022-05-20 |
| 0dd6813 | jens-daniel-mueller | 2022-05-15 |
| 06054b2 | jens-daniel-mueller | 2022-05-10 |
[1] "NorESM-OC1.2_3D_ALL_v20211125"
[1] "dissic" "no3" "o2" "po4" "si" "so" "talk" "thetao"
[1] "dissic_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
dissic lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 2.17 184. -78.5 0
2 2.17 184. -78.5 0
3 2.15 186. -78.5 0
4 2.15 186. -78.5 0
5 2.14 188. -78.5 0
6 2.14 188. -78.5 0
7 2.13 190. -78.5 0
8 2.13 190. -78.5 0
9 2.12 192. -78.5 0
10 2.12 192. -78.5 0
# … with 2,360,490 more rows
[1] "no3_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
no3 lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 0.0199 184. -78.5 0
2 0.0198 184. -78.5 0
3 0.0201 186. -78.5 0
4 0.0205 186. -78.5 0
5 0.0208 188. -78.5 0
6 0.0210 188. -78.5 0
7 0.0210 190. -78.5 0
8 0.0210 190. -78.5 0
9 0.0209 192. -78.5 0
10 0.0208 192. -78.5 0
# … with 2,360,490 more rows
[1] "o2_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
o2 lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 0.379 184. -78.5 0
2 0.381 184. -78.5 0
3 0.378 186. -78.5 0
4 0.375 186. -78.5 0
5 0.371 188. -78.5 0
6 0.368 188. -78.5 0
7 0.367 190. -78.5 0
8 0.366 190. -78.5 0
9 0.365 192. -78.5 0
10 0.364 192. -78.5 0
# … with 2,360,490 more rows
[1] "po4_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
po4 lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 0.00135 184. -78.5 0
2 0.00135 184. -78.5 0
3 0.00136 186. -78.5 0
4 0.00139 186. -78.5 0
5 0.00141 188. -78.5 0
6 0.00142 188. -78.5 0
7 0.00142 190. -78.5 0
8 0.00142 190. -78.5 0
9 0.00141 192. -78.5 0
10 0.00140 192. -78.5 0
# … with 2,360,490 more rows
[1] "si_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
si lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 0.0352 184. -78.5 0
2 0.0354 184. -78.5 0
3 0.0355 186. -78.5 0
4 0.0355 186. -78.5 0
5 0.0354 188. -78.5 0
6 0.0353 188. -78.5 0
7 0.0352 190. -78.5 0
8 0.0351 190. -78.5 0
9 0.0350 192. -78.5 0
10 0.0348 192. -78.5 0
# … with 2,360,490 more rows
[1] "so_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
so lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 32.8 184. -78.5 0
2 32.8 184. -78.5 0
3 32.7 186. -78.5 0
4 32.7 186. -78.5 0
5 32.6 188. -78.5 0
6 32.6 188. -78.5 0
7 32.5 190. -78.5 0
8 32.5 190. -78.5 0
9 32.5 192. -78.5 0
10 32.4 192. -78.5 0
# … with 2,360,490 more rows
[1] "talk_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
talk lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 2.31 184. -78.5 0
2 2.30 184. -78.5 0
3 2.28 186. -78.5 0
4 2.27 186. -78.5 0
5 2.26 188. -78.5 0
6 2.26 188. -78.5 0
7 2.25 190. -78.5 0
8 2.24 190. -78.5 0
9 2.24 192. -78.5 0
10 2.23 192. -78.5 0
# … with 2,360,490 more rows
[1] "thetao_NorESM-OC1.2_A_1_gr_1980-2018_v20211125.nc"
# A tibble: 2,360,500 × 4
thetao lon lat depth
<dbl> <dbl> <dbl> <dbl>
1 -1.17 184. -78.5 0
2 -1.21 184. -78.5 0
3 -1.34 186. -78.5 0
4 -1.43 186. -78.5 0
5 -1.48 188. -78.5 0
6 -1.52 188. -78.5 0
7 -1.56 190. -78.5 0
8 -1.59 190. -78.5 0
9 -1.62 192. -78.5 0
10 -1.64 192. -78.5 0
# … with 2,360,490 more rows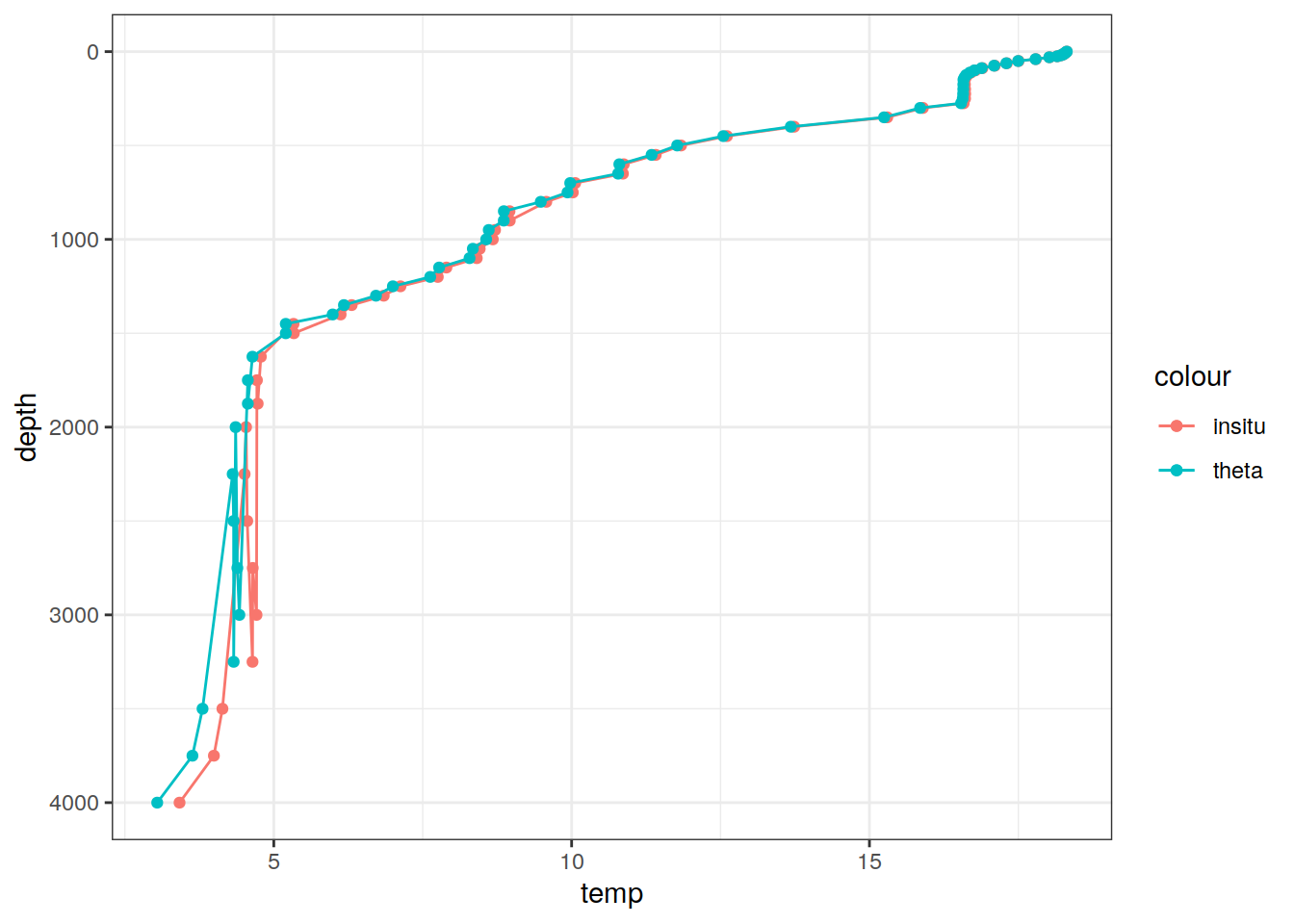
[1] "ORCA1-LIM3-PISCES_3D_ALL_v20211215"
[1] "dissic" "no3" "o2" "po4" "si" "so" "talk" "thetao"
[1] "dissic_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
dissic 0.6356738 2.047529 2.106403 2.107058 2.175867 2.607979 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "no3_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
no3 3.094286e-07 2.920019e-05 0.003260584 0.00814951 0.01467556 0.03648942
NA's
no3 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "o2_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
o2 0.1961508 0.2091869 0.2505452 0.2691502 0.3308462 0.436595 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "po4_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu.
po4 6.226775e-07 0.0001672927 0.0004754509 0.0007070058 0.001179957
Max. NA's
po4 0.008807107 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "si_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
si 0.0008841127 0.001731399 0.003098852 0.01157017 0.01246982 0.08248688 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "so_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
so 5.547729 33.98625 34.35794 34.32555 35.19701 38.63853 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "talk_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
talk 0.6077311 2.330598 2.354492 2.350225 2.383013 2.943625 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229)
[1] "thetao_ORCA1-LIM3-PISCES_A_1_gr_1980-2018_v20211215.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
thetao -1.902045 2.372617 16.12284 14.44789 25.67046 30.61959 30015
dimension(s):
from to offset delta refsys point values x/y
lon 1 360 0 1 WGS 84 NA NULL [x]
lat 1 180 -90 1 WGS 84 NA NULL [y]
depth 1 75 NA NA NA FALSE [0,1.023907),...,[5800,6004.229) 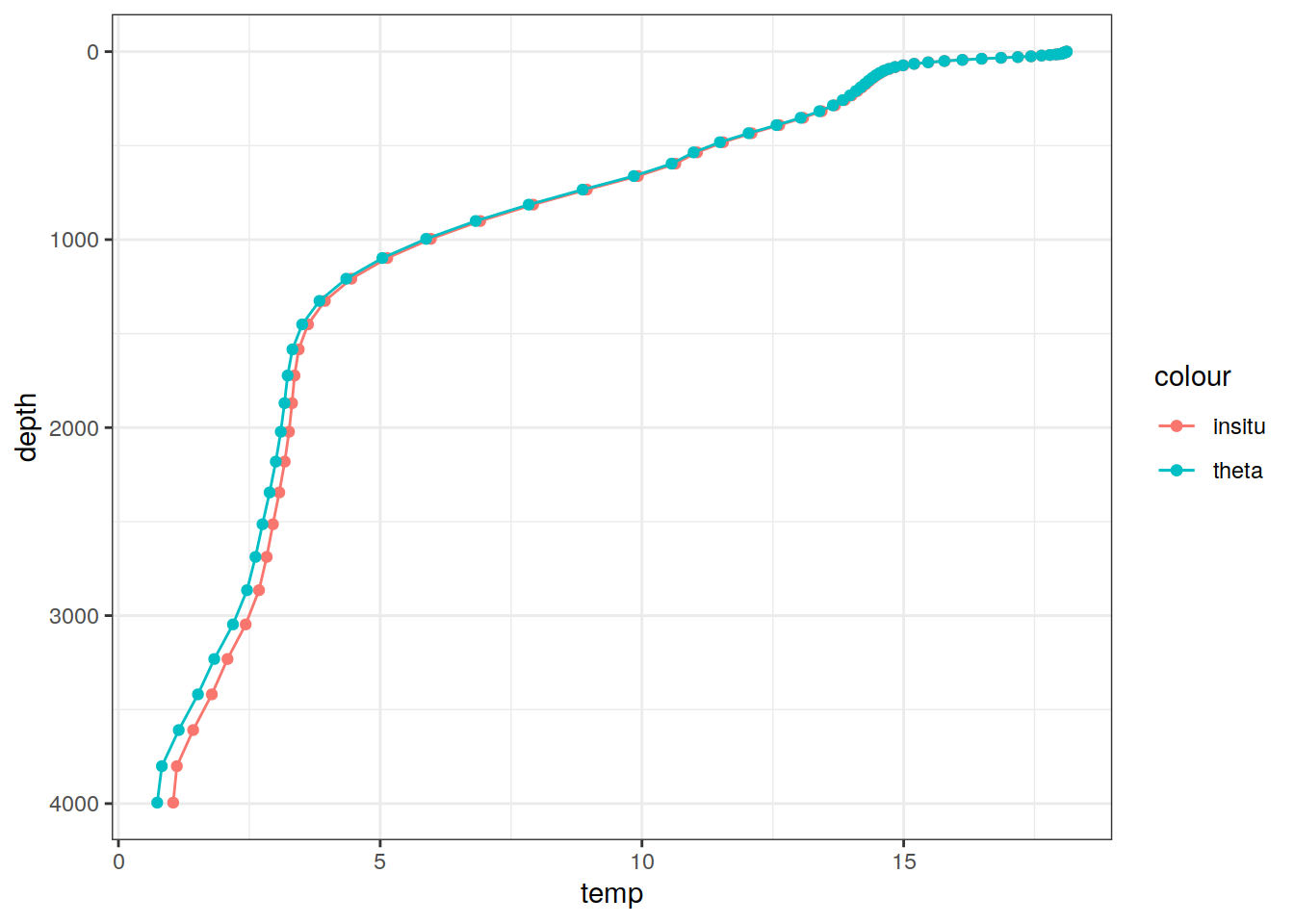
[1] "planktom12_3d_all_v20220404"
[1] "dissic" "o2" "po4" "si" "so" "talk" "thetao"
[1] "dissic_PlankTOM12_A_1_gr_1980-2018_v20220404.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
dissic 0.3604033 1.986821 2.02884 2.063821 2.127698 2.448838 34691
dimension(s):
from to offset delta refsys point values x/y
LONGITUDE 1 360 0 1 NA NA NULL [x]
LATITUDE 1 180 -90 1 NA NA NULL [y]
DEPTH 1 31 NA NA NA NA [31] 4.99994,...,5250.23
[1] "o2_PlankTOM12_A_1_gr_1980-2018_v20220404.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
o2 0.06550782 0.2083643 0.2492491 0.2661756 0.3294328 0.3942035 34691
dimension(s):
from to offset delta refsys point values x/y
LONGITUDE 1 360 0 1 NA NA NULL [x]
LATITUDE 1 180 -90 1 NA NA NULL [y]
DEPTH 1 31 NA NA NA NA [31] 4.99994,...,5250.23
[1] "po4_PlankTOM12_A_1_gr_1980-2018_v20220404.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
po4 0.001492676 0.0351725 0.09724818 0.1070409 0.166172 0.2820027 34691
dimension(s):
from to offset delta refsys point values x/y
LONGITUDE 1 360 0 1 NA NA NULL [x]
LATITUDE 1 180 -90 1 NA NA NULL [y]
DEPTH 1 31 NA NA NA NA [31] 4.99994,...,5250.23
[1] "si_PlankTOM12_A_1_gr_1980-2018_v20220404.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
si 1.032564e-05 3.253466e-05 7.616759e-05 0.01748056 0.006404537 0.1061623
NA's
si 34691
dimension(s):
from to offset delta refsys point values x/y
LONGITUDE 1 360 0 1 NA NA NULL [x]
LATITUDE 1 180 -90 1 NA NA NULL [y]
DEPTH 1 31 NA NA NA NA [31] 4.99994,...,5250.23
[1] "so_PlankTOM12_A_1_gr_1980-2018_v20220404.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
so 1.501021 33.65757 34.32087 34.22928 36.15146 42.97074 34691
dimension(s):
from to offset delta refsys point values x/y
LONGITUDE 1 360 0 1 NA NA NULL [x]
LATITUDE 1 180 -90 1 NA NA NULL [y]
DEPTH 1 31 NA NA NA NA [31] 4.99994,...,5250.23
[1] "talk_PlankTOM12_A_1_gr_1980-2018_v20220404.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
talk 0.3749107 2.286771 2.331598 2.313961 2.375072 3.040825 34691
dimension(s):
from to offset delta refsys point values x/y
LONGITUDE 1 360 0 1 NA NA NULL [x]
LATITUDE 1 180 -90 1 NA NA NULL [y]
DEPTH 1 31 NA NA NA NA [31] 4.99994,...,5250.23
[1] "thetao_PlankTOM12_A_1_gr_1980-2018_v20220404.nc"
stars object with 3 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
thetao -1.839474 2.963888 16.64129 14.84514 25.90274 32.04654 34691
dimension(s):
from to offset delta refsys point values x/y
LONGITUDE 1 360 0 1 NA NA NULL [x]
LATITUDE 1 180 -90 1 NA NA NULL [y]
DEPTH 1 31 NA NA NA NA [31] 4.99994,...,5250.23 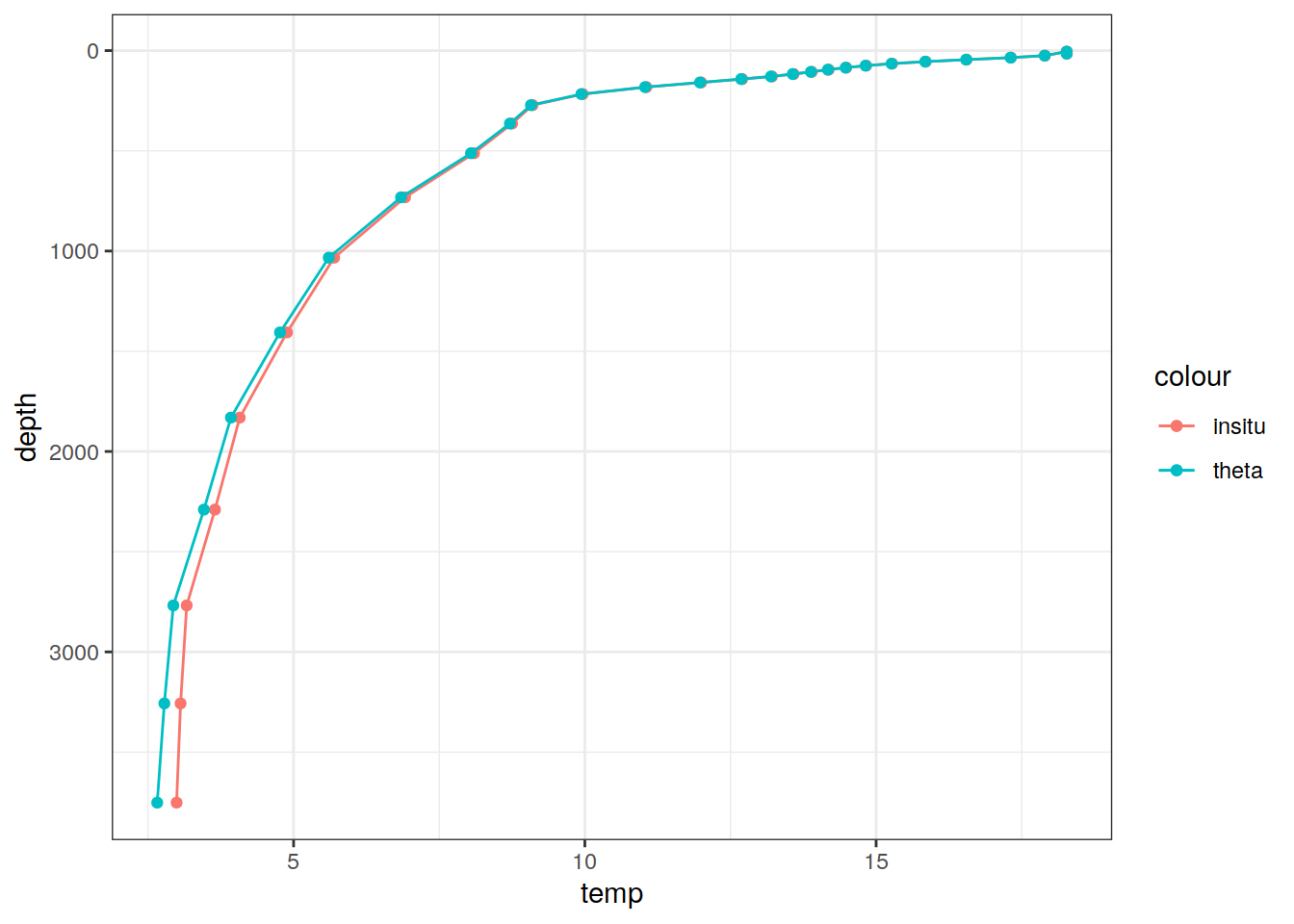
3 Plots
Below, following subsets of the climatology are plotted for all relevant predictors:
- Horizontal planes at 4 depth levels
- Global section along track indicated by white lines in maps.
climatology_files <- fs::dir_ls(path_preprocessing)
climatology <- climatology_files %>%
map_dfr(read_csv, .id = "model")
climatology <- climatology %>%
mutate(model = str_remove(model, path_preprocessing),
model = str_remove(model, "_climatology_runA_2007.csv"))# define plotting variables
vars <- c(
"tco2",
"talk",
"sal",
"nitrate",
"phosphate",
"silicate",
"oxygen",
"temp",
"AOU",
"gamma"
)
# i_var <- vars[1]
for (i_var in vars) {
# i_var <- vars[1]
# plot sections
print(p_section_global(df = climatology %>%
filter(model == models[1]),
var = i_var))
}
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.3
Matrix products: default
BLAS: /usr/local/R-4.1.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.1.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] geosphere_1.5-14 oce_1.5-0 gsw_1.0-6 reticulate_1.23
[5] lubridate_1.8.0 stars_0.5-5 sf_1.0-5 abind_1.4-5
[9] geomtextpath_0.1.0 colorspace_2.0-2 marelac_2.1.10 shape_1.4.6
[13] ggforce_0.3.3 metR_0.11.0 scico_1.3.0 patchwork_1.1.1
[17] collapse_1.7.0 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[21] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tibble_3.1.6
[25] ggplot2_3.3.5 tidyverse_1.3.1 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] ellipsis_0.3.2 class_7.3-20 rprojroot_2.0.2 fs_1.5.2
[5] rstudioapi_0.13 proxy_0.4-26 farver_2.1.0 bit64_4.0.5
[9] fansi_1.0.2 xml2_1.3.3 ncdf4_1.19 knitr_1.37
[13] polyclip_1.10-0 jsonlite_1.7.3 broom_0.7.11 dbplyr_2.1.1
[17] png_0.1-7 compiler_4.1.2 httr_1.4.2 backports_1.4.1
[21] assertthat_0.2.1 Matrix_1.4-0 fastmap_1.1.0 cli_3.1.1
[25] later_1.3.0 tweenr_1.0.2 htmltools_0.5.2 tools_4.1.2
[29] gtable_0.3.0 glue_1.6.0 rappdirs_0.3.3 Rcpp_1.0.8
[33] RNetCDF_2.5-2 cellranger_1.1.0 jquerylib_0.1.4 vctrs_0.3.8
[37] PCICt_0.5-4.1 lwgeom_0.2-8 xfun_0.29 ps_1.6.0
[41] rvest_1.0.2 ncmeta_0.3.0 lifecycle_1.0.1 tidync_0.2.4
[45] getPass_0.2-2 MASS_7.3-55 scales_1.1.1 vroom_1.5.7
[49] hms_1.1.1 promises_1.2.0.1 parallel_4.1.2 yaml_2.2.1
[53] sass_0.4.0 stringi_1.7.6 highr_0.9 e1071_1.7-9
[57] checkmate_2.0.0 rlang_1.0.2 pkgconfig_2.0.3 systemfonts_1.0.3
[61] evaluate_0.14 lattice_0.20-45 SolveSAPHE_2.1.0 labeling_0.4.2
[65] bit_4.0.4 processx_3.5.2 tidyselect_1.1.1 here_1.0.1
[69] seacarb_3.3.0 magrittr_2.0.1 R6_2.5.1 generics_0.1.1
[73] DBI_1.1.2 pillar_1.6.4 haven_2.4.3 whisker_0.4
[77] withr_2.4.3 units_0.7-2 sp_1.4-6 modelr_0.1.8
[81] crayon_1.4.2 KernSmooth_2.23-20 utf8_1.2.2 tzdb_0.2.0
[85] rmarkdown_2.11 grid_4.1.2 readxl_1.3.1 data.table_1.14.2
[89] callr_3.7.0 git2r_0.29.0 reprex_2.0.1 digest_0.6.29
[93] classInt_0.4-3 httpuv_1.6.5 textshaping_0.3.6 munsell_0.5.0
[97] bslib_0.3.1