NIES-ML3_GCB
Jens Daniel Müller
06 March, 2025
Last updated: 2025-03-06
Checks: 7 0
Knit directory:
heatwave_co2_flux_2023/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240307) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8936cec. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data
Ignored: output/
Untracked files:
Untracked: analysis/CSIR-ML6_GCB.Rmd
Untracked: code/gas_flux_kwko.R
Unstaged changes:
Modified: analysis/_site.yml
Modified: analysis/child/pCO2_product_analysis.Rmd
Modified: analysis/child/pCO2_product_preprocessing.Rmd
Modified: analysis/child/pCO2_product_synopsis.Rmd
Modified: code/Workflowr_project_managment.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/NIES-ML3_GCB.Rmd) and HTML
(docs/NIES-ML3_GCB.html) files. If you’ve configured a
remote Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8936cec | jens-daniel-mueller | 2025-03-06 | Include Watson product |
center <- -160
boundary <- center + 180
target_crs <- paste0("+proj=robin +over +lon_0=", center)
# target_crs <- paste0("+proj=eqearth +over +lon_0=", center)
# target_crs <- paste0("+proj=eqearth +lon_0=", center)
# target_crs <- paste0("+proj=igh_o +lon_0=", center)
worldmap <- ne_countries(scale = 'small',
type = 'map_units',
returnclass = 'sf')
worldmap <- worldmap %>% st_break_antimeridian(lon_0 = center)
worldmap_trans <- st_transform(worldmap, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans)
coastline <- ne_coastline(scale = 'small', returnclass = "sf")
coastline <- st_break_antimeridian(coastline, lon_0 = 200)
coastline_trans <- st_transform(coastline, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col="grey") +
# geom_sf(data = coastline_trans)
bbox <- st_bbox(c(xmin = -180, xmax = 180, ymax = 65, ymin = -78), crs = st_crs(4326))
bbox <- st_as_sfc(bbox)
bbox_trans <- st_break_antimeridian(bbox, lon_0 = center)
bbox_graticules <- st_graticule(
x = bbox_trans,
crs = st_crs(bbox_trans),
datum = st_crs(bbox_trans),
lon = c(20, 20.001),
lat = c(-78,65),
ndiscr = 1e3,
margin = 0.001
)
bbox_graticules_trans <- st_transform(bbox_graticules, crs = target_crs)
rm(worldmap, coastline, bbox, bbox_trans)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col="grey") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans)
lat_lim <- ext(bbox_graticules_trans)[c(3,4)]*1.002
lon_lim <- ext(bbox_graticules_trans)[c(1,2)]*1.005
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey90", col = "grey90") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans, linewidth = 1) +
# coord_sf(crs = target_crs,
# ylim = lat_lim,
# xlim = lon_lim,
# expand = FALSE) +
# theme(
# panel.border = element_blank(),
# axis.text = element_blank(),
# axis.ticks = element_blank()
# )
latitude_graticules <- st_graticule(
x = bbox_graticules,
crs = st_crs(bbox_graticules),
datum = st_crs(bbox_graticules),
lon = c(20, 20.001),
lat = c(-60,-30,0,30,60),
ndiscr = 1e3,
margin = 0.001
)
latitude_graticules_trans <- st_transform(latitude_graticules, crs = target_crs)
latitude_labels <- data.frame(lat_label = c("60°N","30°N","Eq.","30°S","60°S"),
lat = c(60,30,0,-30,-60)-4, lon = c(35)-c(0,2,4,2,0))
latitude_labels <- st_as_sf(x = latitude_labels,
coords = c("lon", "lat"),
crs = "+proj=longlat")
latitude_labels_trans <- st_transform(latitude_labels, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col = "grey") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans) +
# geom_sf(data = latitude_graticules_trans,
# col = "grey60",
# linewidth = 0.2) +
# geom_sf_text(data = latitude_labels_trans,
# aes(label = lat_label),
# size = 3,
# col = "grey60")Read data
path_pCO2_products <-
"/nfs/kryo/work/datasets/gridded/ocean/2d/observation/pco2/gcb_2024_pco2_products/"library(ncdf4)
nc <-
nc_open(paste0(
path_pCO2_products,
"GCB-2024_dataprod_NIES-ML3_1982-2023.nc"
))
print(nc)pco2_product <-
read_ncdf(
paste0(
path_pCO2_products,
"GCB-2024_dataprod_NIES-ML3_1982-2023.nc"
),
var = c("fco2atm", "fgco2", "kw", "alpha", "sfco2", "tos"),
ignore_bounds = TRUE,
make_units = FALSE
)
pco2_product <- pco2_product %>%
as_tibble()
pco2_product <- pco2_product %>%
drop_na()
pco2_product <-
pco2_product %>%
rename(temperature = tos,
atm_fco2 = fco2atm,
sol = alpha)
pco2_product <-
pco2_product %>%
mutate(area = earth_surf(lat, lon),
year = year(time),
month = month(time))
pco2_product <-
pco2_product %>%
mutate(lon = if_else(lon < 20, lon + 360, lon))
pco2_product <-
pco2_product %>%
mutate(dfco2 = sfco2 - atm_fco2)
pco2_product <-
pco2_product %>%
mutate(fgco2 = -fgco2 * 60 * 60 * 24 * 365,
kw_sol = kw * sol * 1e-2 * 24 * 365)
pco2_product <-
pco2_product %>%
select(-c(kw, sol))pCO2_product_preprocessing <-
knitr::knit_expand(
file = here::here("analysis/child/pCO2_product_preprocessing.Rmd"),
product_name = "NIES-ML3_GCB"
)Preprocessing
# model <- TRUE
model <- str_detect('NIES-ML3_GCB', "FESOM-REcoM|ETHZ-CESM")Load masks
biome_mask <-
read_rds(here::here("data/biome_mask.rds"))
region_mask <-
read_rds(here::here("data/region_mask.rds"))
map <-
read_rds(here::here("data/map.rds"))
key_biomes <-
read_rds(here::here("data/key_biomes.rds"))Define labels and breaks
labels_breaks <- function(i_name) {
if (i_name == "dco2") {
i_legend_title <- "ΔpCO<sub>2</sub><br>(µatm)"
}
if (i_name == "dfco2") {
i_legend_title <- "ΔfCO<sub>2</sub><br>(µatm)"
}
if (i_name == "atm_co2") {
i_legend_title <- "pCO<sub>2,atm</sub><br>(µatm)"
}
if (i_name == "atm_fco2") {
i_legend_title <- "fCO<sub>2,atm</sub><br>(µatm)"
}
if (i_name == "sol") {
i_legend_title <- "K<sub>0</sub><br>(mol m<sup>-3</sup> µatm<sup>-1</sup>)"
}
if (i_name == "kw") {
i_legend_title <- "k<sub>w</sub><br>(m yr<sup>-1</sup>)"
}
if (i_name == "kw_sol") {
i_legend_title <- "k<sub>w</sub> K<sub>0</sub><br>(mol yr<sup>-1</sup> m<sup>-2</sup> µatm<sup>-1</sup>)"
}
if (i_name == "spco2") {
i_legend_title <- "pCO<sub>2,ocean</sub><br>(µatm)"
}
if (i_name == "sfco2") {
i_legend_title <- "fCO<sub>2,ocean</sub><br>(µatm)"
}
if (i_name == "intpp") {
i_legend_title <- "NPP<sub>int</sub><br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
}
if (i_name == "no3") {
i_legend_title <- "NO<sub>3</sub><br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "o2") {
i_legend_title <- "O<sub>2</sub><br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "dissic") {
i_legend_title <- "DIC<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "sdissic") {
i_legend_title <- "sDIC<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "cstar") {
i_legend_title <- "C*<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "talk") {
i_legend_title <- "TA<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "stalk") {
i_legend_title <- "sTA<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "sdissic_stalk") {
i_legend_title <- "sDIC-sTA<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "sfco2_total") {
i_legend_title <- "total"
}
if (i_name == "sfco2_therm") {
i_legend_title <- "thermal"
}
if (i_name == "sfco2_nontherm") {
i_legend_title <- "non-thermal"
}
if (i_name == "fgco2") {
i_legend_title <- "FCO<sub>2</sub><br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
}
if (i_name == "fgco2_predict") {
i_legend_title <- "FCO<sub>2</sub> pred.<br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
}
if (i_name == "fgco2_hov") {
i_legend_title <- "FCO<sub>2</sub><br>(PgC deg<sup>-1</sup> yr<sup>-1</sup>)"
}
if (i_name == "fgco2_int") {
i_legend_title <- "FCO<sub>2</sub><br>(PgC yr<sup>-1</sup>)"
}
if (i_name == "fgco2_predict_int") {
i_legend_title <- "FCO<sub>2</sub> pred.<br>(PgC yr<sup>-1</sup>)"
}
if (i_name == "thetao") {
i_legend_title <- "Temp.<br>(°C)"
}
if (i_name == "temperature") {
i_legend_title <- "SST<br>(°C)"
}
if (i_name == "salinity") {
i_legend_title <- "SSS"
}
if (i_name == "so") {
i_legend_title <- "salinity"
}
if (i_name == "chl") {
i_legend_title <- "lg(Chl-a)<br>(lg(mg m<sup>-3</sup>))"
}
if (i_name == "mld") {
i_legend_title <- "MLD<br>(m)"
}
if (i_name == "press") {
i_legend_title <- "pressure<sub>atm</sub><br>(Pa)"
}
if (i_name == "wind") {
i_legend_title <- "Wind <br>(m sec<sup>-1</sup>)"
}
if (i_name == "SSH") {
i_legend_title <- "SSH <br>(m)"
}
if (i_name == "fice") {
i_legend_title <- "Sea ice <br>(%)"
}
if (i_name == "resid_fgco2") {
i_legend_title <-
"Observed"
}
if (i_name == "resid_fgco2_dfco2") {
i_legend_title <-
"ΔfCO<sub>2</sub>"
}
if (i_name == "resid_fgco2_kw_sol") {
i_legend_title <-
"k<sub>w</sub> K<sub>0</sub>"
}
if (i_name == "resid_fgco2_dfco2_kw_sol") {
i_legend_title <-
"k<sub>w</sub> K<sub>0</sub> X ΔfCO<sub>2</sub>"
}
if (i_name == "resid_fgco2_sum") {
i_legend_title <-
"∑"
}
if (i_name == "resid_fgco2_offset") {
i_legend_title <-
"Obs. - ∑"
}
all_labels_breaks <- lst(i_legend_title)
return(all_labels_breaks)
}
x_axis_labels <-
c(
"dco2" = labels_breaks("dco2")$i_legend_title,
"dfco2" = labels_breaks("dfco2")$i_legend_title,
"atm_co2" = labels_breaks("atm_co2")$i_legend_title,
"atm_fco2" = labels_breaks("atm_fco2")$i_legend_title,
"sol" = labels_breaks("sol")$i_legend_title,
"kw" = labels_breaks("kw")$i_legend_title,
"kw_sol" = labels_breaks("kw_sol")$i_legend_title,
"intpp" = labels_breaks("intpp")$i_legend_title,
"no3" = labels_breaks("no3")$i_legend_title,
"o2" = labels_breaks("o2")$i_legend_title,
"dissic" = labels_breaks("dissic")$i_legend_title,
"sdissic" = labels_breaks("sdissic")$i_legend_title,
"cstar" = labels_breaks("cstar")$i_legend_title,
"talk" = labels_breaks("talk")$i_legend_title,
"stalk" = labels_breaks("stalk")$i_legend_title,
"sdissic_stalk" = labels_breaks("sdissic_stalk")$i_legend_title,
"spco2" = labels_breaks("spco2")$i_legend_title,
"sfco2" = labels_breaks("sfco2")$i_legend_title,
"sfco2_total" = labels_breaks("sfco2_total")$i_legend_title,
"sfco2_therm" = labels_breaks("sfco2_therm")$i_legend_title,
"sfco2_nontherm" = labels_breaks("sfco2_nontherm")$i_legend_title,
"fgco2" = labels_breaks("fgco2")$i_legend_title,
"fgco2_predict" = labels_breaks("fgco2_predict")$i_legend_title,
"fgco2_hov" = labels_breaks("fgco2_hov")$i_legend_title,
"fgco2_int" = labels_breaks("fgco2_int")$i_legend_title,
"fgco2_predict_int" = labels_breaks("fgco2_int")$i_legend_title,
"thetao" = labels_breaks("thetao")$i_legend_title,
"temperature" = labels_breaks("temperature")$i_legend_title,
"salinity" = labels_breaks("salinity")$i_legend_title,
"so" = labels_breaks("so")$i_legend_title,
"chl" = labels_breaks("chl")$i_legend_title,
"mld" = labels_breaks("mld")$i_legend_title,
"press" = labels_breaks("press")$i_legend_title,
"wind" = labels_breaks("wind")$i_legend_title,
"SSH" = labels_breaks("SSH")$i_legend_title,
"fice" = labels_breaks("fice")$i_legend_title,
"resid_fgco2" = labels_breaks("resid_fgco2")$i_legend_title,
"resid_fgco2_dfco2" = labels_breaks("resid_fgco2_dfco2")$i_legend_title,
"resid_fgco2_kw_sol" = labels_breaks("resid_fgco2_kw_sol")$i_legend_title,
"resid_fgco2_dfco2_kw_sol" = labels_breaks("resid_fgco2_dfco2_kw_sol")$i_legend_title,
"resid_fgco2_sum" = labels_breaks("resid_fgco2_sum")$i_legend_title,
"resid_fgco2_offset" = labels_breaks("resid_fgco2_offset")$i_legend_title
)Analysis settings
name_quadratic_fit <- c("atm_co2", "atm_fco2", "spco2", "sfco2")
# name_quadratic_fit <- c(name_quadratic_fit, "dfco2", "fgco2", "fgco2_int", "temperature")
start_year <- 1990
name_divergent <- c("dco2", "dfco2", "fgco2", "fgco2_hov", "fgco2_int")Data preprocessing
pco2_product <-
pco2_product %>%
filter(year >= start_year)pco2_product_interior <-
pco2_product_interior %>%
filter(time >= ymd(paste0(start_year, "-01-01")))biome_mask <- biome_mask %>%
mutate(area = earth_surf(lat, lon))
pco2_product <-
full_join(pco2_product,
biome_mask)
# set all values outside biome mask to NA
pco2_product <-
pco2_product %>%
mutate(across(-c(lat, lon, time, area, year, month, biome),
~ if_else(is.na(biome), NA, .)))Compuations
Maps
Biome means
pco2_product_biome_monthly_global <-
pco2_product %>%
filter(!is.na(fgco2)) %>%
mutate(fgco2_int = fgco2) %>%
mutate(biome = case_when(str_detect(biome, "SO-SPSS|SO-ICE|Arctic") ~ "Polar",
TRUE ~ "Global non-polar")) %>%
filter(biome == "Global non-polar") %>%
select(-c(lon, lat, year, month)) %>%
group_by(time, biome) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_monthly_biome <-
pco2_product %>%
filter(!is.na(fgco2)) %>%
mutate(fgco2_int = fgco2) %>%
select(-c(lon, lat, year, month)) %>%
group_by(time, biome) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_monthly <-
bind_rows(pco2_product_biome_monthly_global,
pco2_product_biome_monthly_biome)
rm(
pco2_product_biome_monthly_global,
pco2_product_biome_monthly_biome
)
pco2_product_biome_monthly <-
pco2_product_biome_monthly %>%
filter(!is.na(biome))
pco2_product_biome_monthly <-
pco2_product_biome_monthly %>%
mutate(year = year(time),
month = month(time),
.after = time)
pco2_product_biome_monthly <-
pco2_product_biome_monthly %>%
pivot_longer(-c(time, year, month, biome))
pco2_product_biome_annual <-
pco2_product_biome_monthly %>%
group_by(year, biome, name) %>%
summarise(value = mean(value)) %>%
ungroup()Profiles
pco2_product_interior <-
left_join(
biome_mask,
pco2_product_interior
)
pco2_product_profiles <- pco2_product_interior %>%
fselect(-c(lat, lon)) %>%
fgroup_by(biome, depth, time) %>% {
add_vars(fgroup_vars(., "unique"),
fmean(.,
w = area,
keep.w = FALSE,
keep.group_vars = FALSE))
}
pco2_product_profiles <-
pco2_product_profiles %>%
mutate(
year = year(time),
month = month(time)
)
gc()Zonal mean sections
pco2_product_interior <-
left_join(
region_mask,
pco2_product_interior %>% select(-c(biome, area))
)
pco2_product_zonal_mean <- pco2_product_interior %>%
fselect(-c(lon)) %>%
fgroup_by(region, depth, lat, time) %>% {
add_vars(fgroup_vars(., "unique"),
fmean(.,
keep.group_vars = FALSE))
}
pco2_product_zonal_mean <-
pco2_product_zonal_mean %>%
mutate(
year = year(time),
month = month(time)
)
gc()
rm(pco2_product_interior)
gc()Absolute values
Hovmoeller plots
The following Hovmoeller plots show the value of each variable as provided through the pCO2 product. Hovmoeller plots are first presented as annual means, and than as monthly means.
Annual means
pco2_product_hovmoeller_annual <-
pco2_product %>%
mutate(fgco2_int = fgco2) %>%
select(-c(lon, time, month, biome)) %>%
group_by(year, lat) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup() %>%
rename(fgco2_hov = fgco2_int) %>%
filter(fgco2_hov != 0)
pco2_product_hovmoeller_annual <-
pco2_product_hovmoeller_annual %>%
pivot_longer(-c(year, lat)) %>%
drop_na()
# pco2_product_hovmoeller_annual %>%
# filter(!(name %in% name_divergent)) %>%
# group_split(name) %>%
# # tail(5) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, lat, fill = value)) +
# geom_raster() +
# scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
# theme(legend.title = element_markdown()) +
# coord_cartesian(expand = 0) +
# labs(title = "Annual means",
# y = "Latitude") +
# theme(axis.title.x = element_blank())
# )
#
# pco2_product_hovmoeller_annual %>%
# filter(name %in% name_divergent) %>%
# group_split(name) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, lat, fill = value)) +
# geom_raster() +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks(.x %>% distinct(name)),
# limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
# oob = squish
# ) +
# theme(legend.title = element_markdown()) +
# coord_cartesian(expand = 0) +
# labs(title = "Annual means",
# y = "Latitude") +
# theme(axis.title.x = element_blank())
# )Monthly means
pco2_product_hovmoeller_monthly <-
pco2_product %>%
mutate(fgco2_int = fgco2) %>%
select(-c(lon, time, biome)) %>%
group_by(year, month, lat) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup() %>%
rename(fgco2_hov = fgco2_int) %>%
filter(fgco2_hov != 0)
pco2_product_hovmoeller_monthly <-
pco2_product_hovmoeller_monthly %>%
pivot_longer(-c(year, month, lat)) %>%
drop_na()
pco2_product_hovmoeller_monthly <-
pco2_product_hovmoeller_monthly %>%
mutate(decimal = year + (month-1) / 12)
# pco2_product_hovmoeller_monthly %>%
# filter(!(name %in% name_divergent)) %>%
# group_split(name) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(decimal, lat, fill = value)) +
# geom_raster() +
# scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
# theme(legend.title = element_markdown()) +
# labs(title = "Monthly means",
# y = "Latitude") +
# coord_cartesian(expand = 0) +
# theme(axis.title.x = element_blank())
# )
#
# pco2_product_hovmoeller_monthly %>%
# filter(name %in% name_divergent) %>%
# group_split(name) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(decimal, lat, fill = value)) +
# geom_raster() +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks(.x %>% distinct(name)),
# limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
# oob = squish
# )+
# theme(legend.title = element_markdown()) +
# labs(title = "Monthly means",
# y = "Latitude") +
# coord_cartesian(expand = 0) +
# theme(axis.title.x = element_blank())
# )rm(pco2_product)
gc()pCO2_product_analysis_2023 <-
knitr::knit_expand(
file = here::here("analysis/child/pCO2_product_analysis.Rmd"),
product_name = "NIES-ML3_GCB",
year_anom = 2023
)2023 anomalies
Functions
Anomaly detection
For the detection of anomalies at any point in time and space, we fit regression models and compare the fitted to the actual value.
We use linear regression models for all parameters, except for , which are approximated with quadratic fits.
The regression models are fitted to all data since , except 2023.
anomaly_determination <- function(df,...) {
group_by <- quos(...)
# group_by <- quos(lon, lat)
# group_by <- quos(biome)
# df <- pco2_product_map_annual
# Linear regression models
df_lm <-
df %>%
filter(year != 2023,
!(name %in% name_quadratic_fit)) %>%
drop_na() %>%
nest(data = -c(name, !!!group_by)) %>%
mutate(fit = map(data, ~ flm(
formula = value ~ year, data = .x
)))
df_lm <-
left_join(
df_lm %>%
unnest_wider(fit) %>%
select(name, !!!group_by,
intercept = `(Intercept)`, slope = year) %>%
mutate(intercept = as.vector(intercept),
slope = as.vector(slope)),
df
) %>%
mutate(fit = intercept + year * slope) %>%
select(name, !!!group_by, year, fit, value) %>%
mutate(resid = value - fit)
# df_lm <-
# df %>%
# filter(year != 2023,
# !(name %in% name_quadratic_fit)) %>%
# drop_na() %>%
# nest(data = -c(name, !!!group_by)) %>%
# mutate(
# fit = map(data, ~ lm(value ~ year, data = .x)),
# tidied = map(fit, tidy),
# augmented = map(fit, augment)
# )
#
#
# df_lm_year_anom <-
# full_join(
# df_lm %>%
# unnest(tidied) %>%
# select(name, !!!group_by, term, estimate) %>%
# pivot_wider(names_from = term,
# values_from = estimate) %>%
# mutate(fit = `(Intercept)` + year * 2023) %>%
# select(name, !!!group_by, fit) %>%
# mutate(year = 2023),
# df %>%
# filter(year == 2023,
# !(name %in% name_quadratic_fit))
# ) %>%
# mutate(resid = value - fit)
#
#
# df_lm <-
# bind_rows(
# df_lm %>%
# unnest(augmented) %>%
# select(name, !!!group_by, year, value, fit = .fitted, resid = .resid),
# df_lm_year_anom
# )
#
# rm(df_lm_year_anom)
# Quadratic regression models
if(any(df %>% distinct(name) %>% pull() %in% name_quadratic_fit)){
df_quadratic <-
df %>%
filter(year != 2023,
name %in% name_quadratic_fit) %>%
drop_na() %>%
mutate(year = year - 2000) %>%
nest(data = -c(name, !!!group_by)) %>%
mutate(
fit = map(data, ~ flm(
# formula = value ~ year + I(year ^ 2), data = .x))
# formula = value ~ year + I(year ^ 2) + I(year ^ 3), data = .x))
formula = value ~ poly(year, 2, raw = TRUE), data = .x))
)
df_quadratic <-
left_join(
df_quadratic %>%
unnest_wider(fit) %>%
select(name, !!!group_by,
intercept = `(Intercept)`,
slope = `1`,
slope_squared = `2`
# slope_cube = `3`
) %>%
mutate(intercept = as.vector(intercept),
slope = as.vector(slope),
slope_squared = as.vector(slope_squared)
# slope_cube = as.vector(slope_cube)
),
df %>%
mutate(year = year - 2000)
) %>%
mutate(fit = intercept + year * slope + year^2 * slope_squared) %>%
select(name, !!!group_by, year, fit, value) %>%
mutate(resid = value - fit,
year = year + 2000)
# df_quadratic <-
# df %>%
# filter(year != 2023,
# name %in% name_quadratic_fit) %>%
# nest(data = -c(name, !!!group_by)) %>%
# mutate(
# fit = map(data, ~ lm(value ~ year + I(year ^ 2), data = .x)),
# tidied = map(fit, tidy),
# augmented = map(fit, augment)
# )
#
# df_quadratic_year_anom <-
# full_join(
# df_quadratic %>%
# unnest(tidied) %>%
# select(name, !!!group_by, term, estimate) %>%
# pivot_wider(names_from = term,
# values_from = estimate) %>%
# mutate(fit = `(Intercept)` + year * 2023 + `I(year^2)` * 2023 ^ 2) %>%
# select(name, !!!group_by, fit) %>%
# mutate(year = 2023),
# df %>%
# filter(year == 2023,
# name %in% name_quadratic_fit)
# ) %>%
# mutate(resid = value - fit)
#
#
# df_quadratic <-
# bind_rows(
# df_quadratic %>%
# unnest(augmented) %>%
# select(name, !!!group_by, year, value, fit = .fitted, resid = .resid),
# df_quadratic_year_anom
# )
#
# rm(df_quadratic_year_anom)
# Join linear and quadratic regression results
df_anomaly <-
bind_rows(df_lm,
df_quadratic)
rm(df_lm,
df_quadratic)
} else{
df_anomaly <- df_lm
rm(df_lm)
}
df_anomaly <-
df_anomaly %>%
arrange(year)
return(df_anomaly)
}Seasonality plots
warm_color <- "#B84A60FF"
cold_color <- "#16877CFF"
p_season <- function(df,
dim_row = "name",
dim_col = "biome",
title = NULL,
var = "resid",
scales = "free_y") {
p <- ggplot(data = df,
aes(month, !!ensym(var)))
if(var == "resid"){
p <- p +
geom_hline(yintercept = 0, linewidth =0.5)
}
p <- p +
geom_path(data = . %>% filter(year != 2023),
aes(group = as.factor(year),
col = as.factor(paste(min(year), max(year), sep = "-"))),
alpha = 0.5)+
geom_path(data = . %>%
filter(year != 2023) %>%
group_by_at(vars(month, dim_col, dim_row)) %>%
summarise(!!ensym(var) := mean(!!ensym(var))),
aes(col = "Climatological\nmean"),
linewidth = 1) +
scale_color_manual(values = c("grey", "black"),
guide = guide_legend(order = 2,
reverse = TRUE)) +
new_scale_color()+
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(
values = warm_color,
guide = guide_legend(order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = title,
x = "Month")
if(df %>% filter(name == "fgco2") %>% nrow() > 0 & "value" %in% names(df)){
df_sink <- df %>%
filter(year == 2023,
name == "fgco2")
p <- p +
geom_point(data = df_sink %>% filter(value < 0),
aes(shape = "Sink"), fill = "white") +
geom_point(data = df_sink %>% filter(value >= 0),
aes(shape = "Source"), fill = "white") +
scale_shape_manual(values = c(25,24))
}
if (!(is.null(dim_col))) {
p <- p +
facet_grid(
as.formula(paste(dim_row, "~", dim_col)),
scales = scales,
labeller = labeller(name = x_axis_labels),
switch = "y"
)
} else {
p <- p +
facet_grid(
as.formula(paste(dim_row, "~ .")),
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
)
}
p <- p +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)
p
}fCO2 decomposition
fco2_decomposition <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
# group_by <- quos(biome, year, month)
pco2_product_biome_monthly_fCO2_decomposition <-
df %>%
filter(name %in% c("temperature", "sfco2"))
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "temperature") %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid),
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "sfco2") %>%
select(-c(value, resid)) %>%
pivot_wider(values_from = fit)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(sfco2_therm = (sfco2 * exp(0.0423 * temperature)) - sfco2)
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition,
df %>%
filter(name %in% c("sfco2")) %>%
select(-c(value, fit, name)) %>%
rename(sfco2_total = resid)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(sfco2_nontherm = sfco2_total - sfco2_therm)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
select(-c(temperature, sfco2)) %>%
pivot_longer(starts_with("sfco2"),
values_to = "resid")
}fco2_decomposition <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
# group_by <- quos(biome, year, month)
pco2_product_biome_monthly_fCO2_decomposition <-
df %>%
filter(name %in% c("temperature", "sfco2"))
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "temperature") %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid),
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "sfco2") %>%
select(-c(value, resid)) %>%
pivot_wider(values_from = fit)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(
sfco2_therm = (sfco2 * exp(0.0423 * temperature)) - sfco2,
sfco2_nontherm = (sfco2 * exp(-0.0423 * temperature)) - sfco2)
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition,
df %>%
filter(name %in% c("sfco2")) %>%
select(-c(value, fit, name)) %>%
rename(sfco2_total = resid)
)
# pco2_product_biome_monthly_fCO2_decomposition <-
# pco2_product_biome_monthly_fCO2_decomposition %>%
# mutate(sfco2_nontherm = sfco2_total - sfco2_therm)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
select(-c(temperature, sfco2)) %>%
pivot_longer(starts_with("sfco2"),
values_to = "resid")
}Flux attribution
flux_attribution <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
pco2_product_flux_attribution <-
df %>%
filter(name %in% c("dfco2", "kw_sol", "fgco2"))
pco2_product_flux_attribution <-
inner_join(
pco2_product_flux_attribution %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid,
names_prefix = "resid_"),
pco2_product_flux_attribution %>%
select(-c(value, resid)) %>%
filter(name != "fgco2") %>%
pivot_wider(values_from = fit)
)
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
mutate(
resid_fgco2_dfco2 = resid_dfco2 * kw_sol,
resid_fgco2_kw_sol = resid_kw_sol * dfco2,
resid_fgco2_dfco2_kw_sol = resid_dfco2 * resid_kw_sol
# resid_fgco2_sum = resid_fgco2_dfco2 + resid_fgco2_kw_sol + resid_fgco2_dfco2_kw_sol
)
# pco2_product_flux_attribution <-
# pco2_product_flux_attribution %>%
# mutate(resid_fgco2_offset = resid_fgco2 - resid_fgco2_sum)
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
select(!!!group_by, starts_with("resid_fgco2")) %>%
pivot_longer(starts_with("resid_"),
values_to = "resid")
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
filter(str_detect(name, "dfco2|kw_sol")) %>%
mutate(name = factor(
name,
levels = c(
"resid_fgco2",
"resid_fgco2_dfco2",
"resid_fgco2_kw_sol",
"resid_fgco2_dfco2_kw_sol",
"resid_fgco2_sum",
"resid_fgco2_offset"
)
))
}Maps
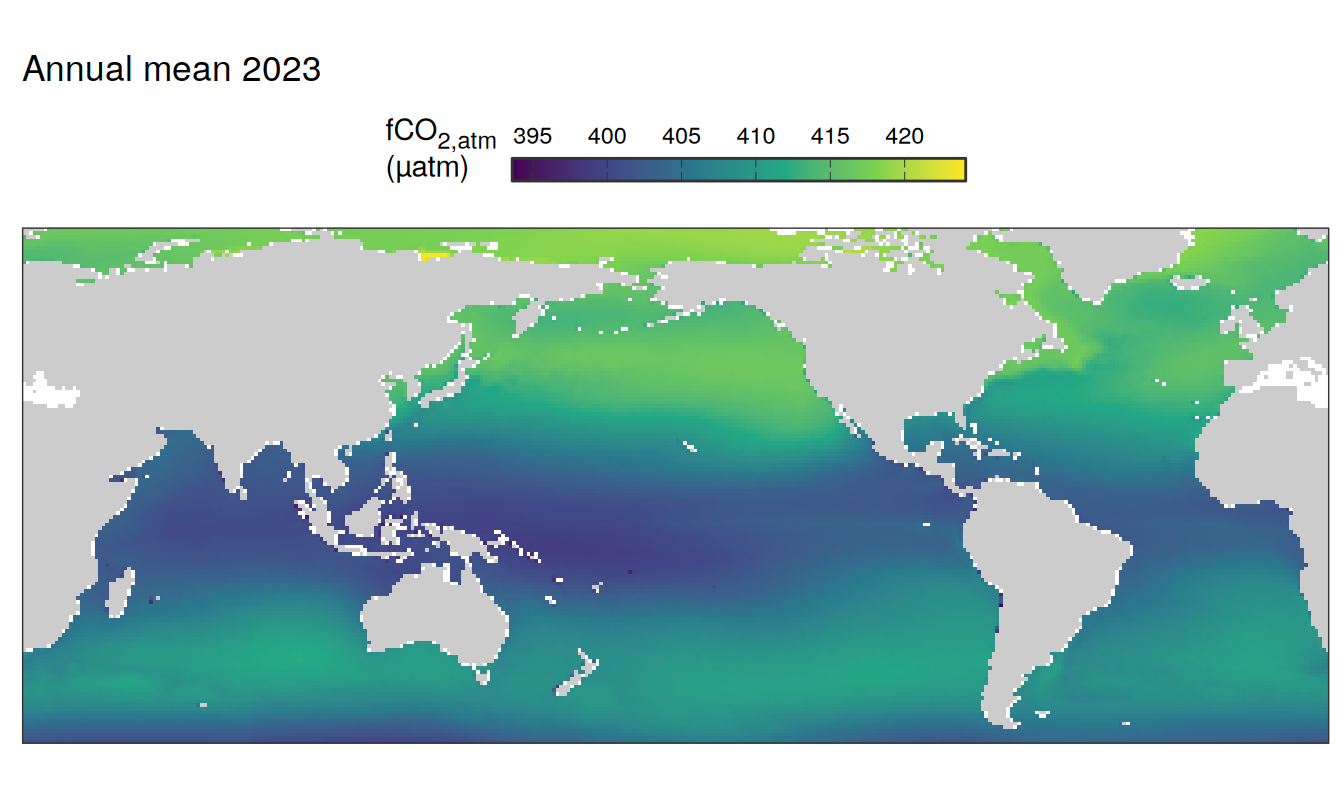
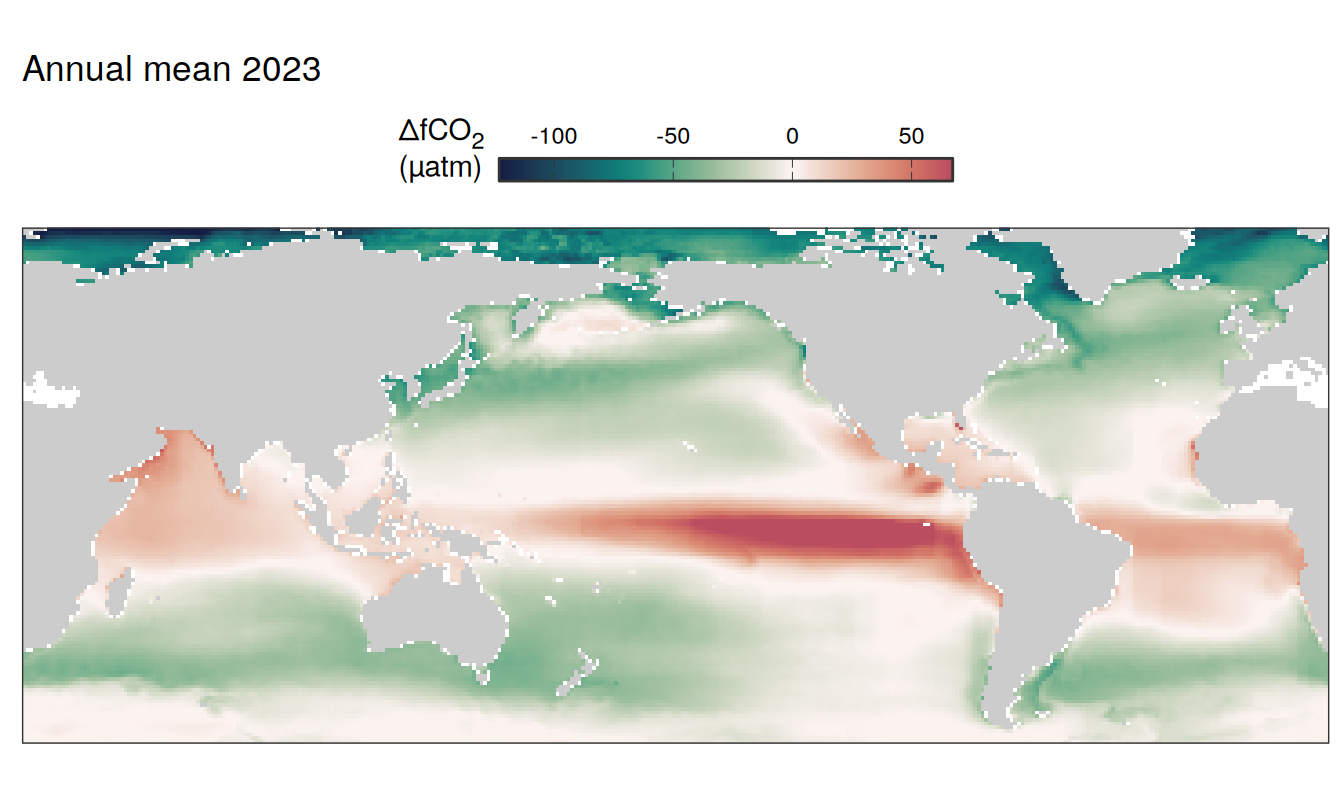
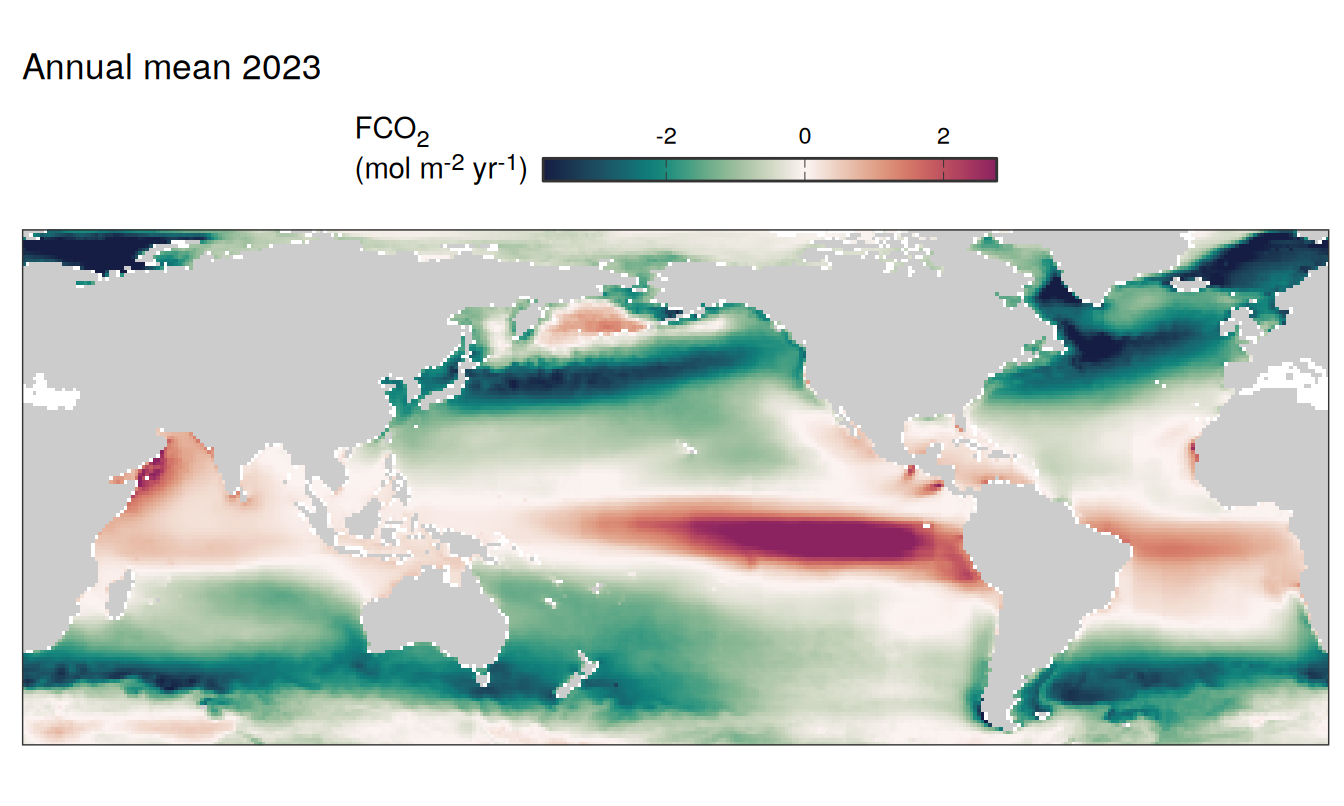
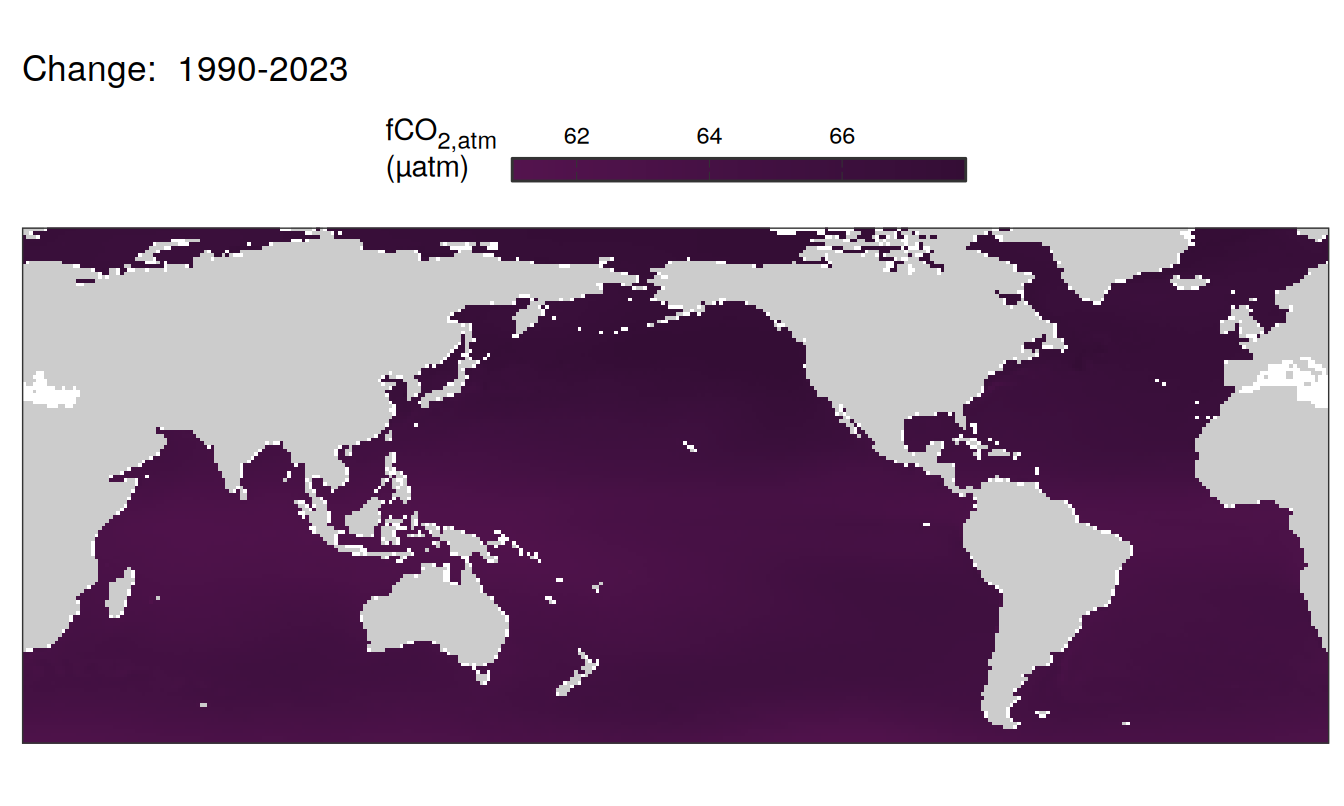
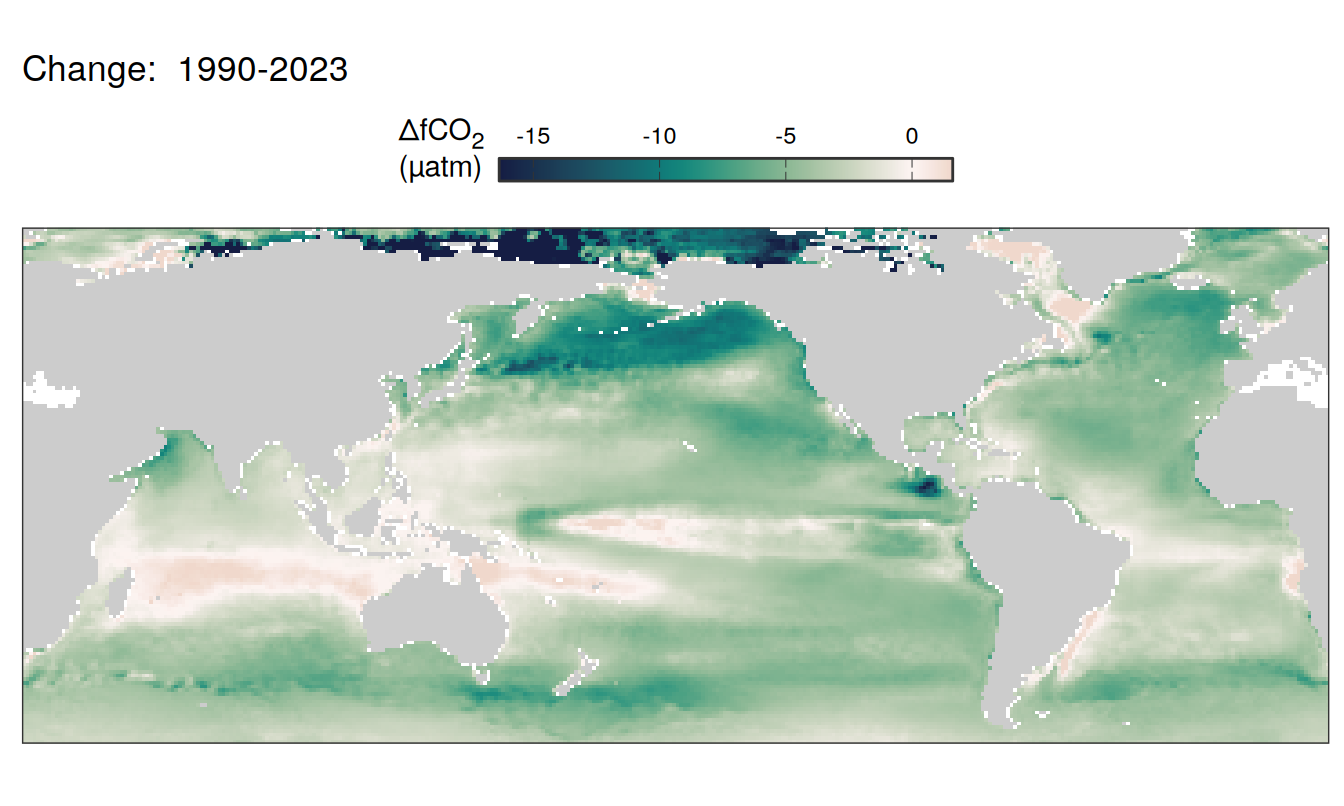
The following maps show the absolute state of each variable in 2023 as provided through the pCO2 product, the change in that variable from 1990 to 2023, as well es the anomalies in 2023. Changes and anomalies are determined based on the predicted value of a linear regression model fit to the data from 1990 to 2022.
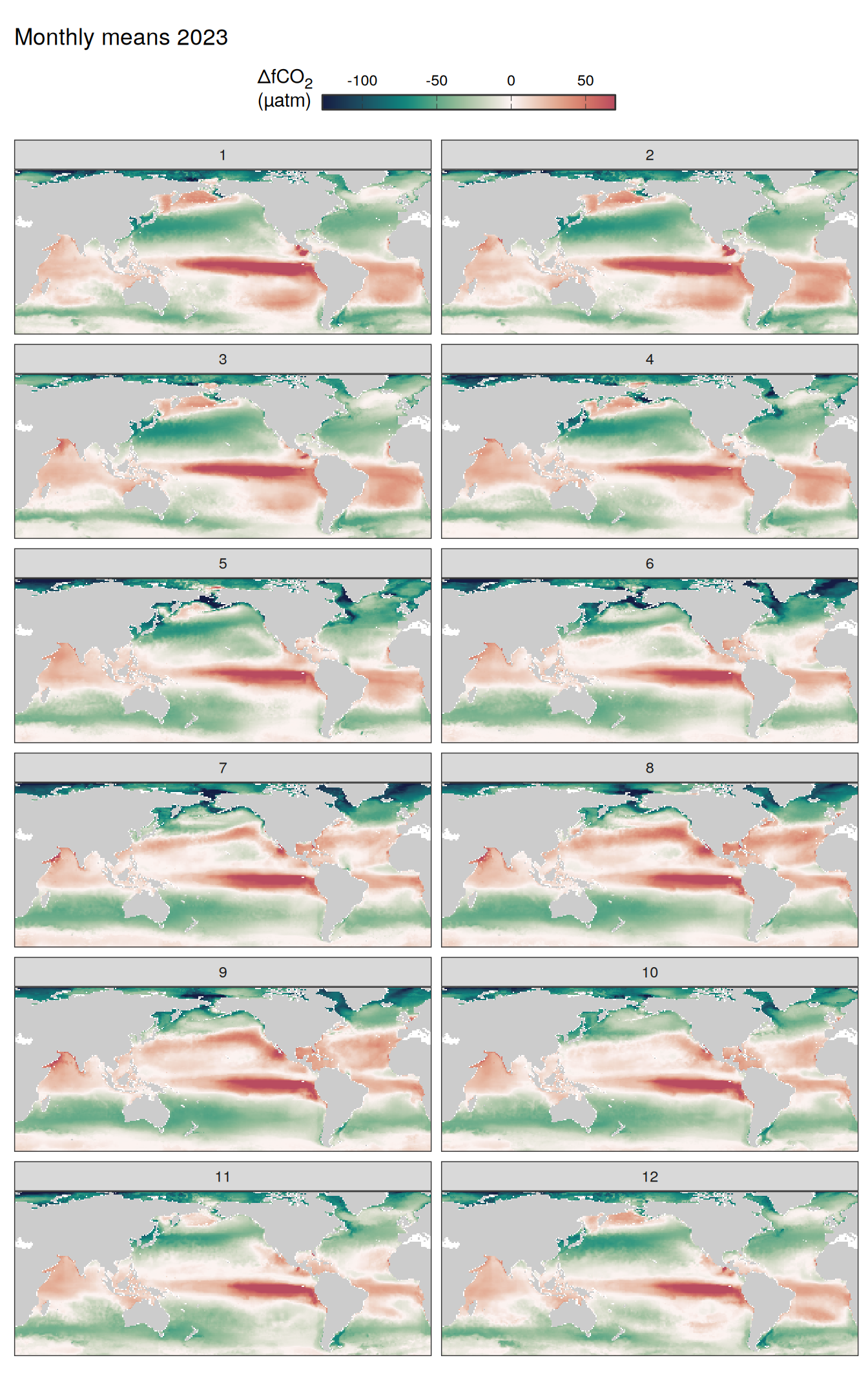
Maps are first presented as annual means, and than as monthly means. Note that the 2023 predictions for the monthly maps are done individually for each month, such the mean seasonal anomaly from the annual mean is removed.
Note: The increase the computational speed, I regridded all maps to 5X5° grid.
Annual means
2023 absolute
pco2_product_map_annual_anomaly <-
pco2_product_map_annual %>%
drop_na() %>%
anomaly_determination(lon, lat)
pco2_product_map_annual_anomaly <-
pco2_product_map_annual_anomaly %>%
drop_na()
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
!(name %in% name_divergent)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = value)) +
labs(title = paste("Annual mean", 2023)) +
scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)
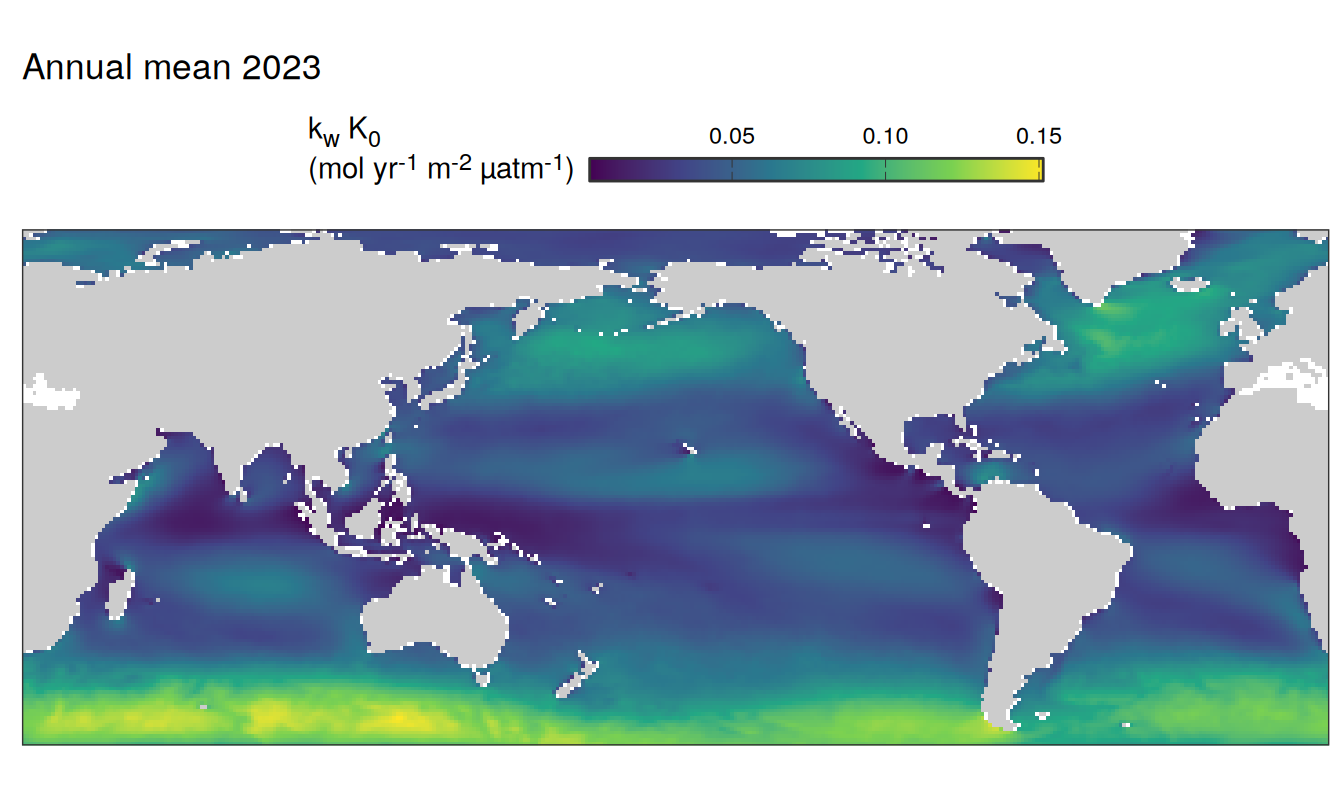
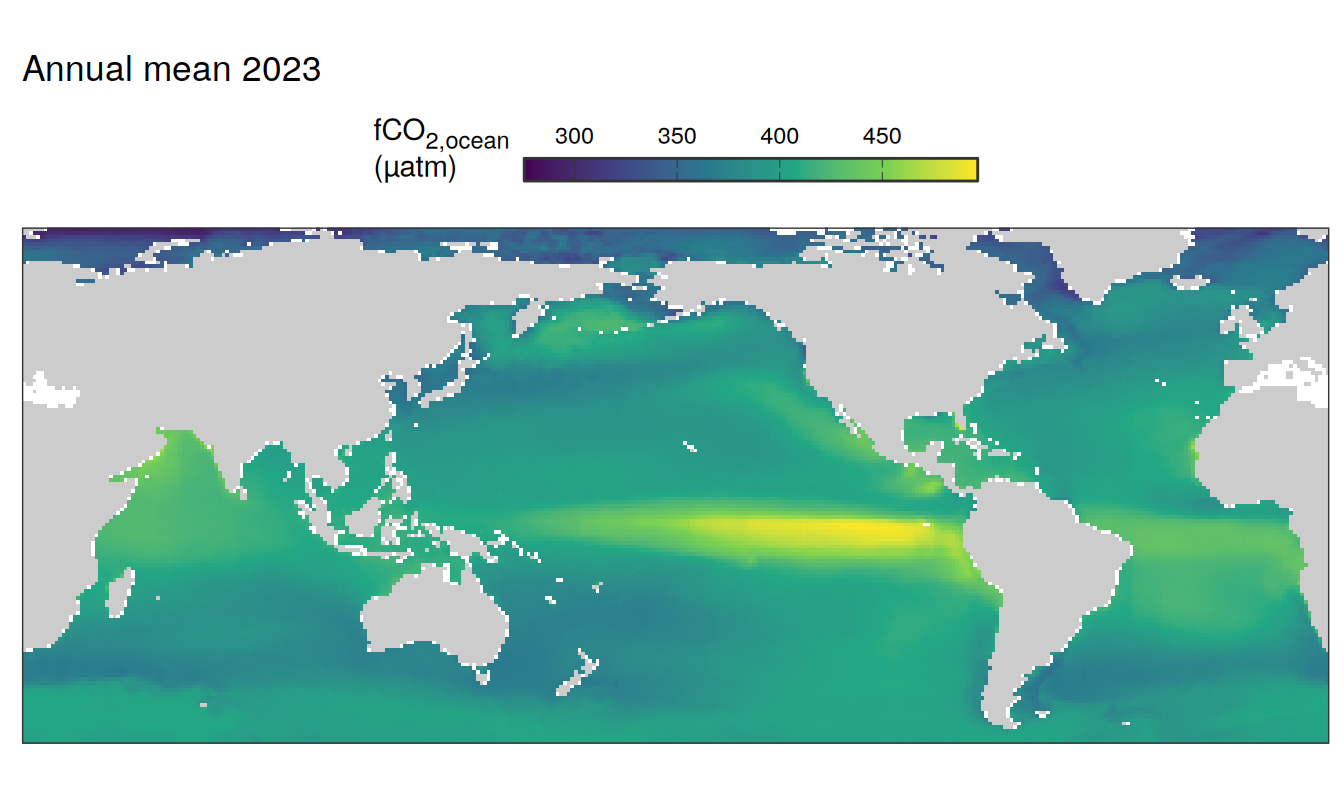
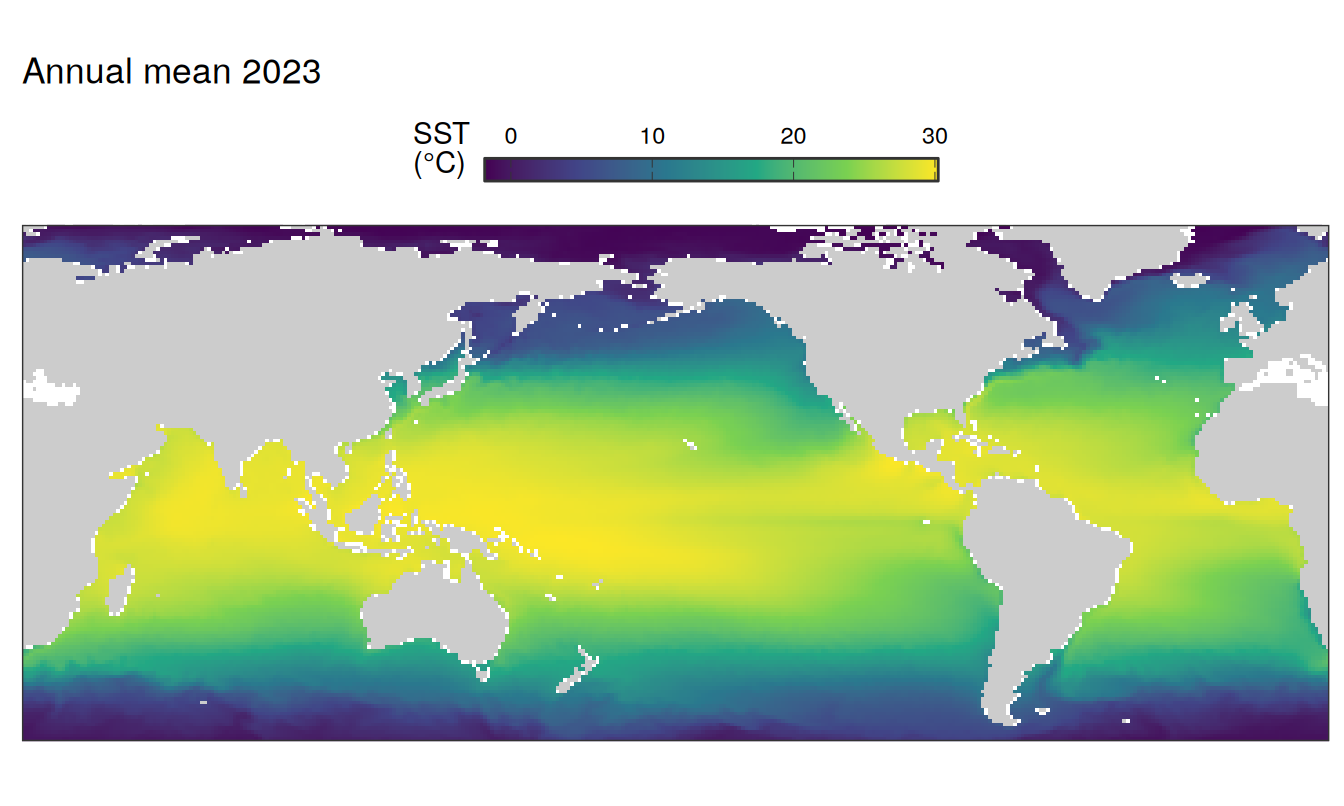
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
name %in% name_divergent) %>%
group_split(name) %>%
# head(1) %>%
map( ~ map +
geom_tile(data = .x,
aes(lon, lat, fill = value)) +
labs(title = paste("Annual mean", 2023)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)
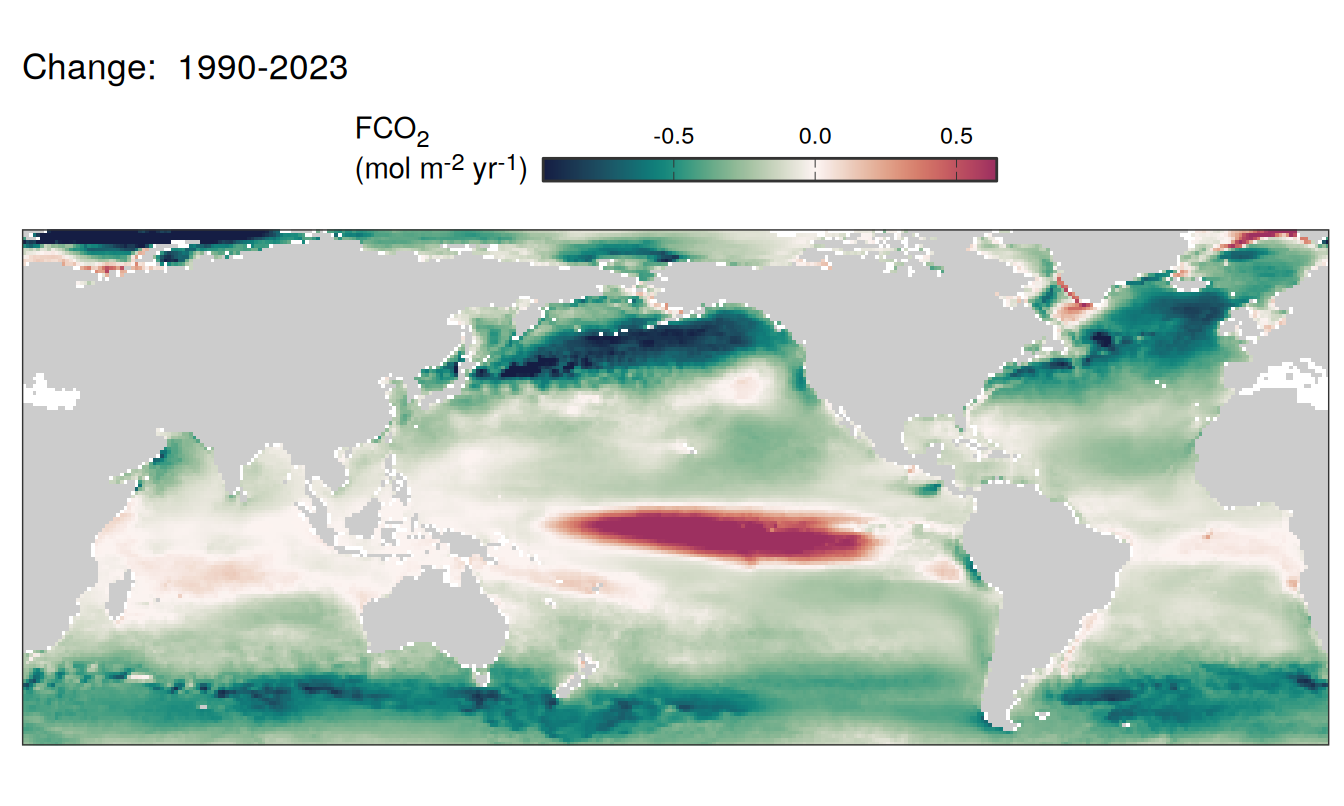
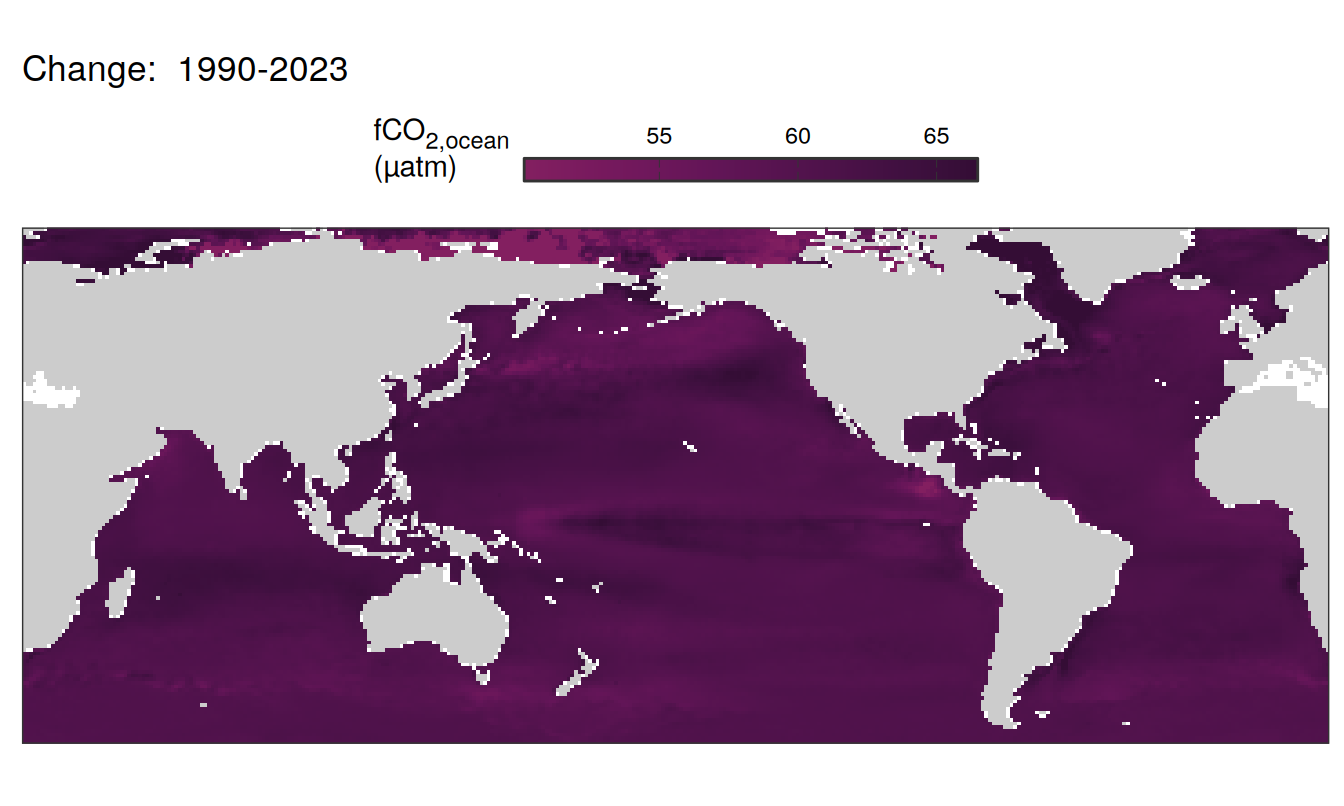
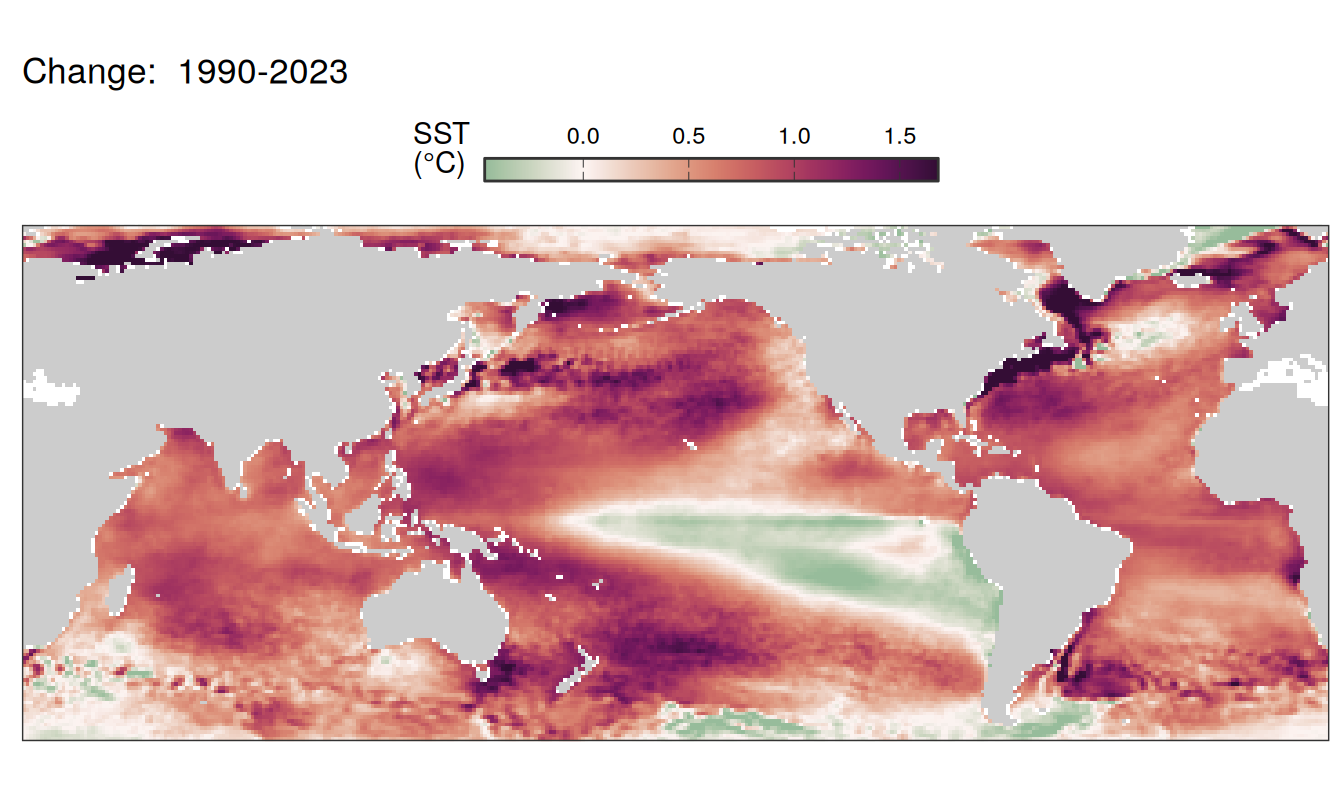
Trends
pco2_product_map_annual_anomaly %>%
group_by(name) %>%
filter(year %in% c(min(year), max(year), 2023)) %>%
ungroup() %>%
select(-c(value, resid)) %>%
filter(year %in% c(min(year), max(year))) %>%
arrange(year) %>%
group_by(lon, lat, name) %>%
mutate(change = fit - lag(fit),
period = paste(lag(year), year, sep = "-")) %>%
ungroup() %>%
filter(!is.na(change)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = change)) +
labs(title = paste("Change: ",.x$period)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$change, .01), quantile(.x$change, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)

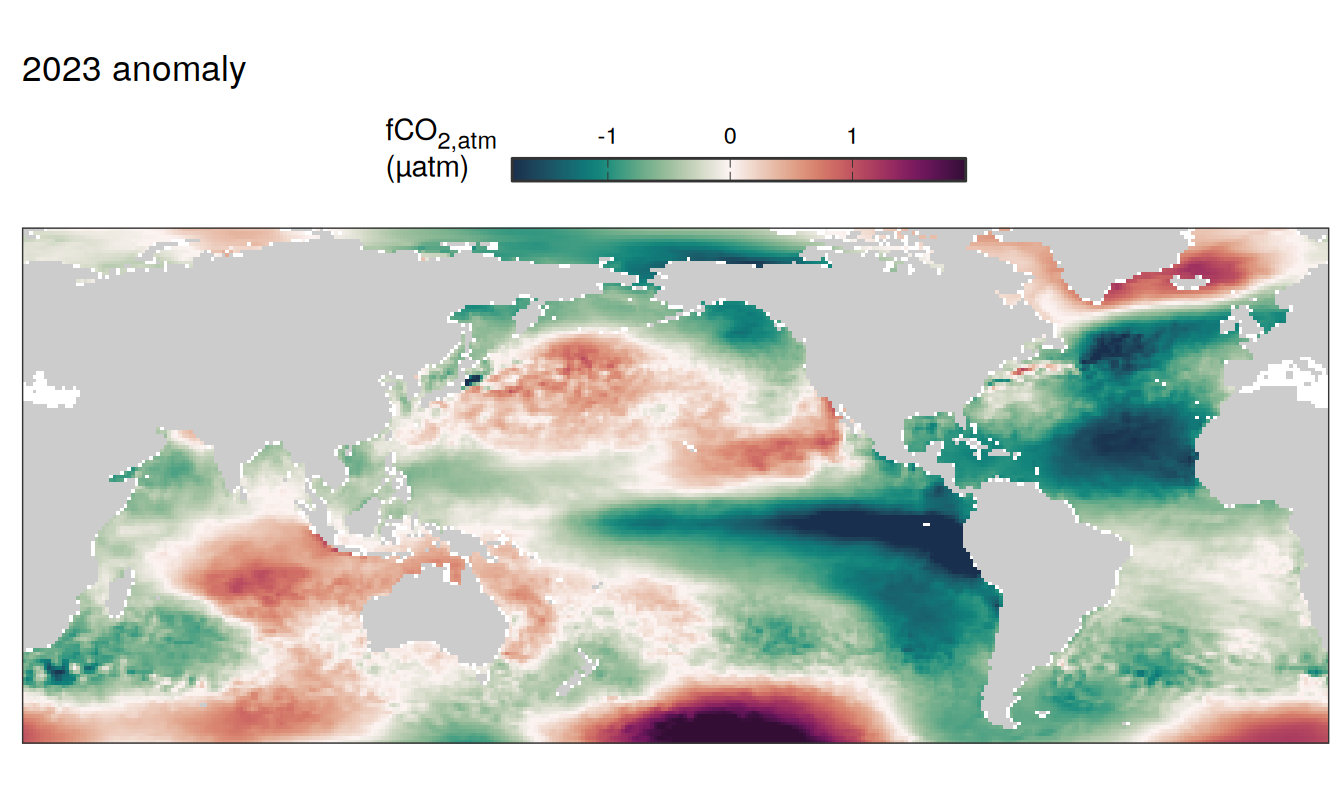
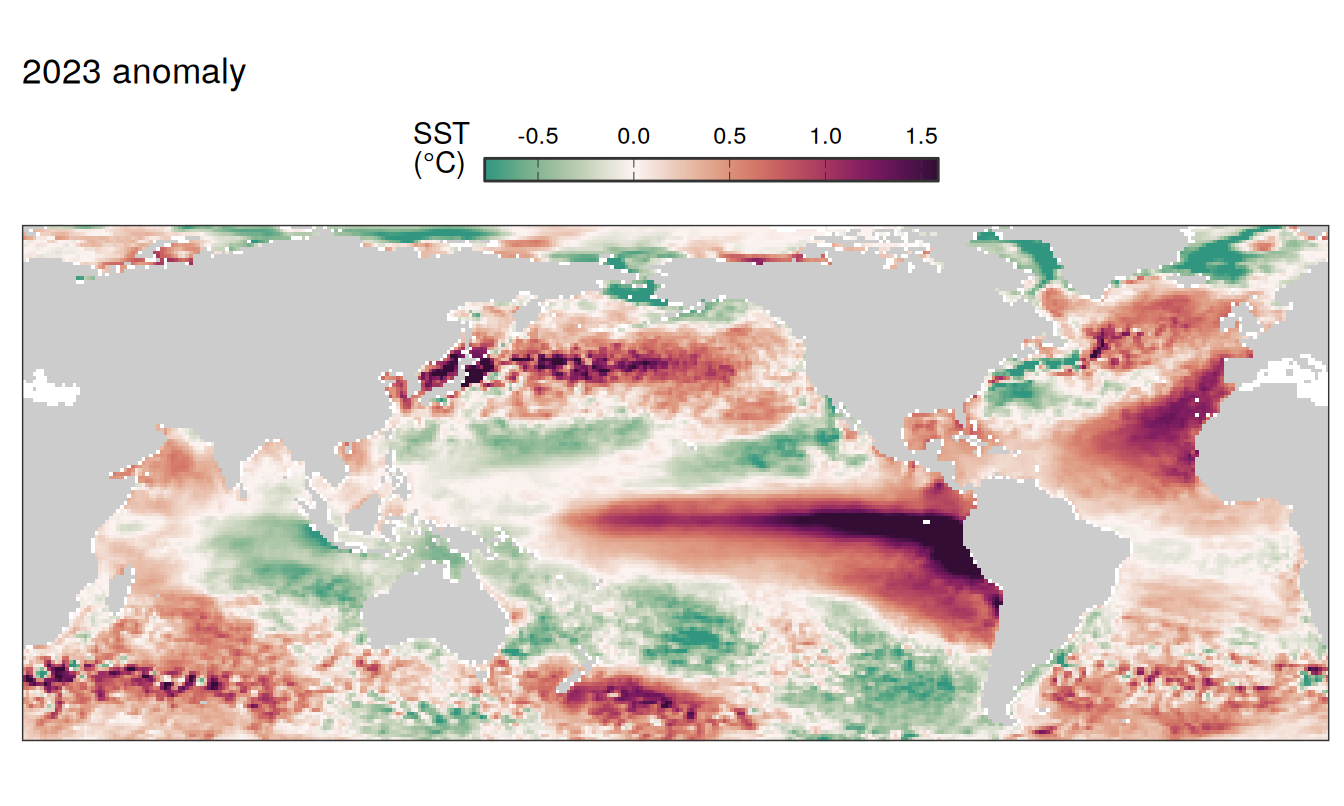
2023 anomaly
pco2_product_map_annual_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(1) %>%
map( ~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid)) +
labs(title = paste(2023,"anomaly")) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
)+
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)
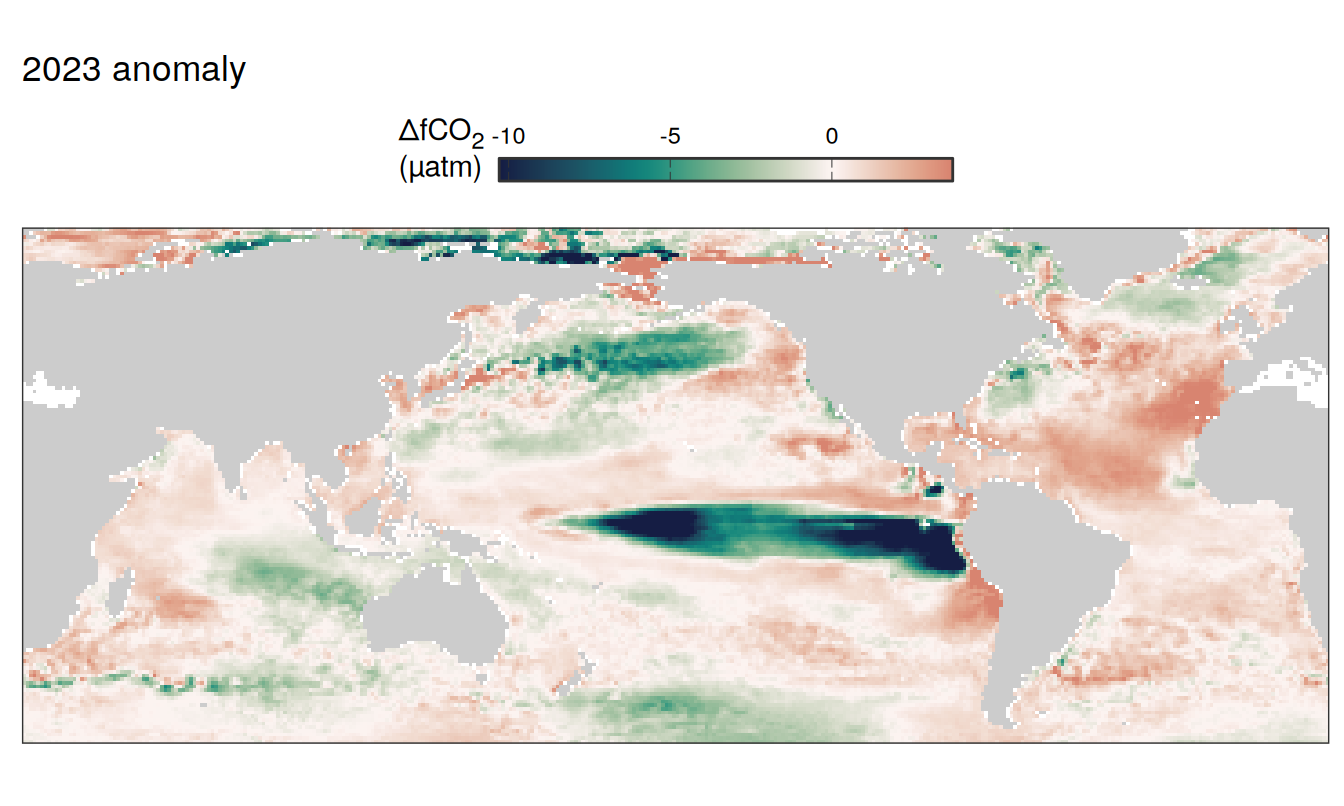
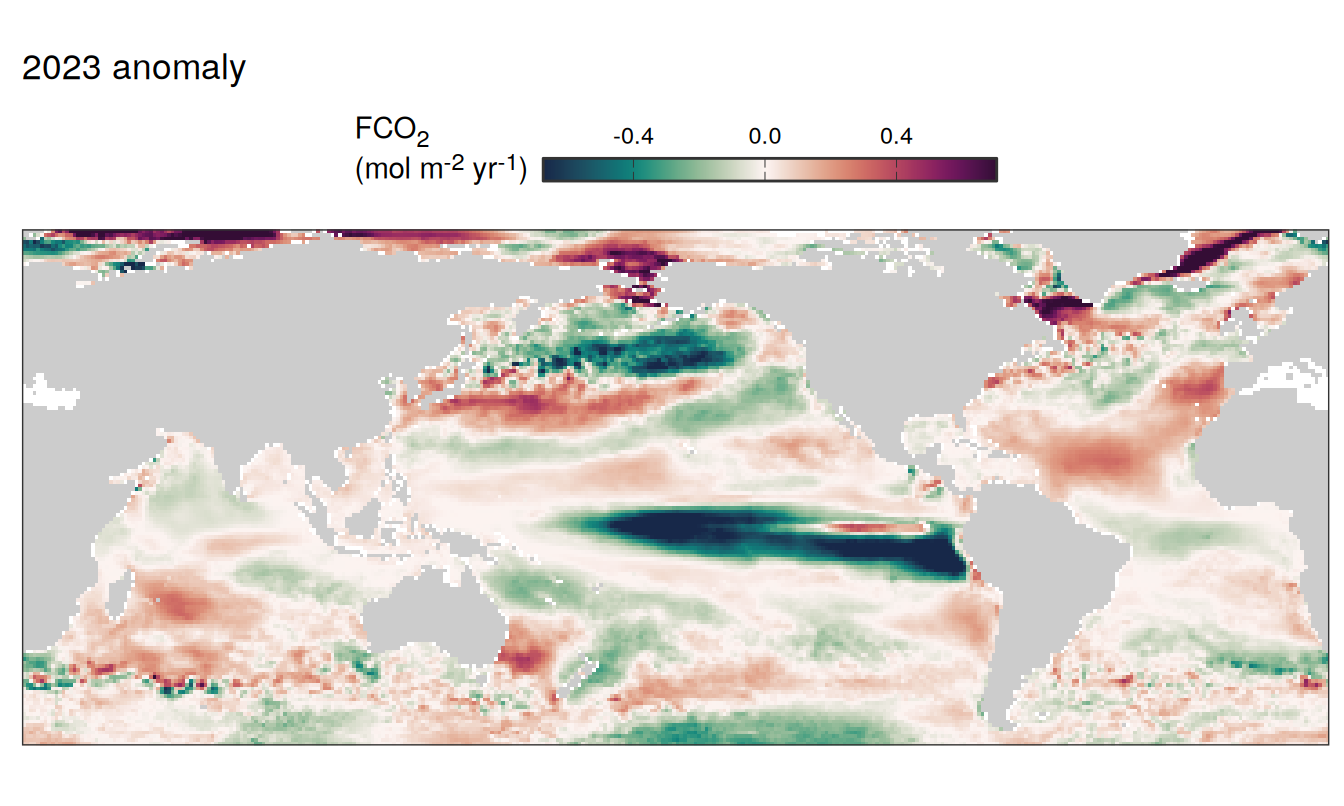
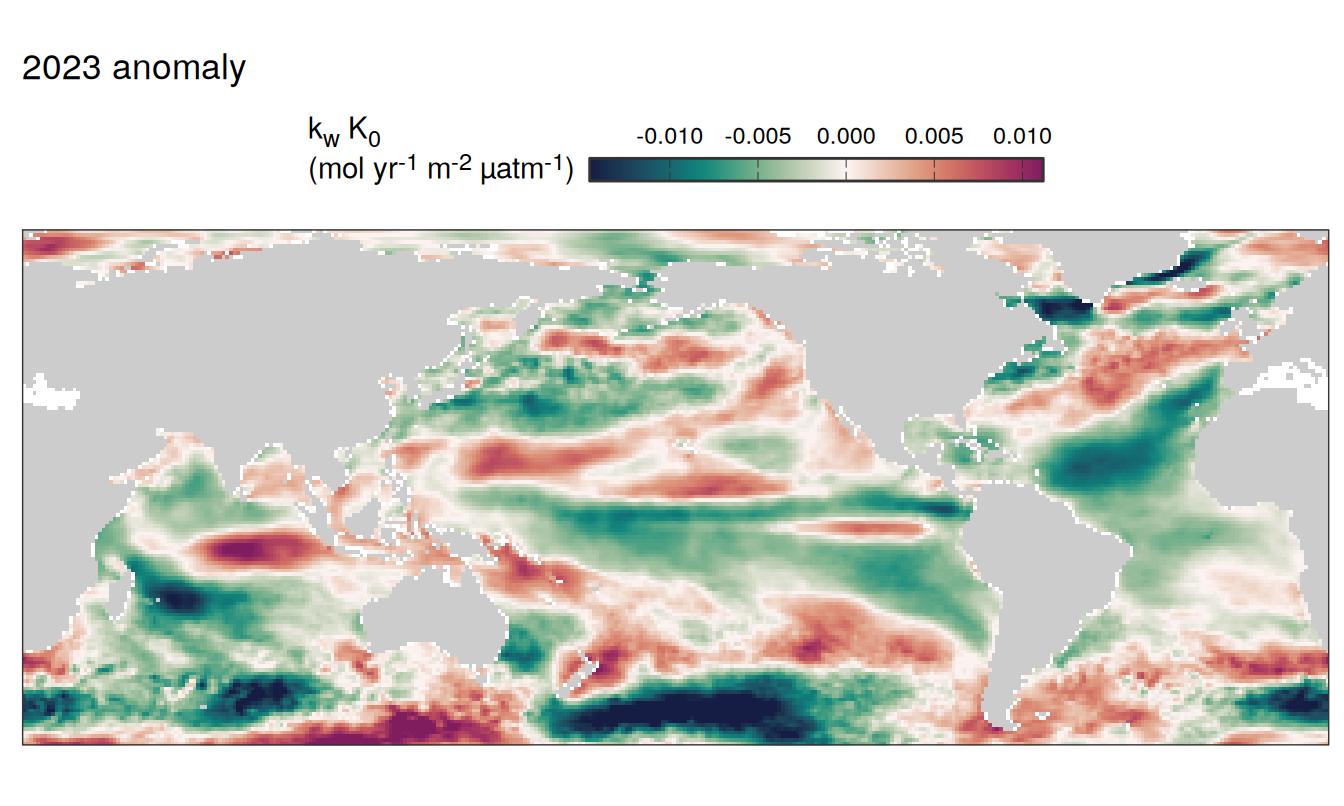
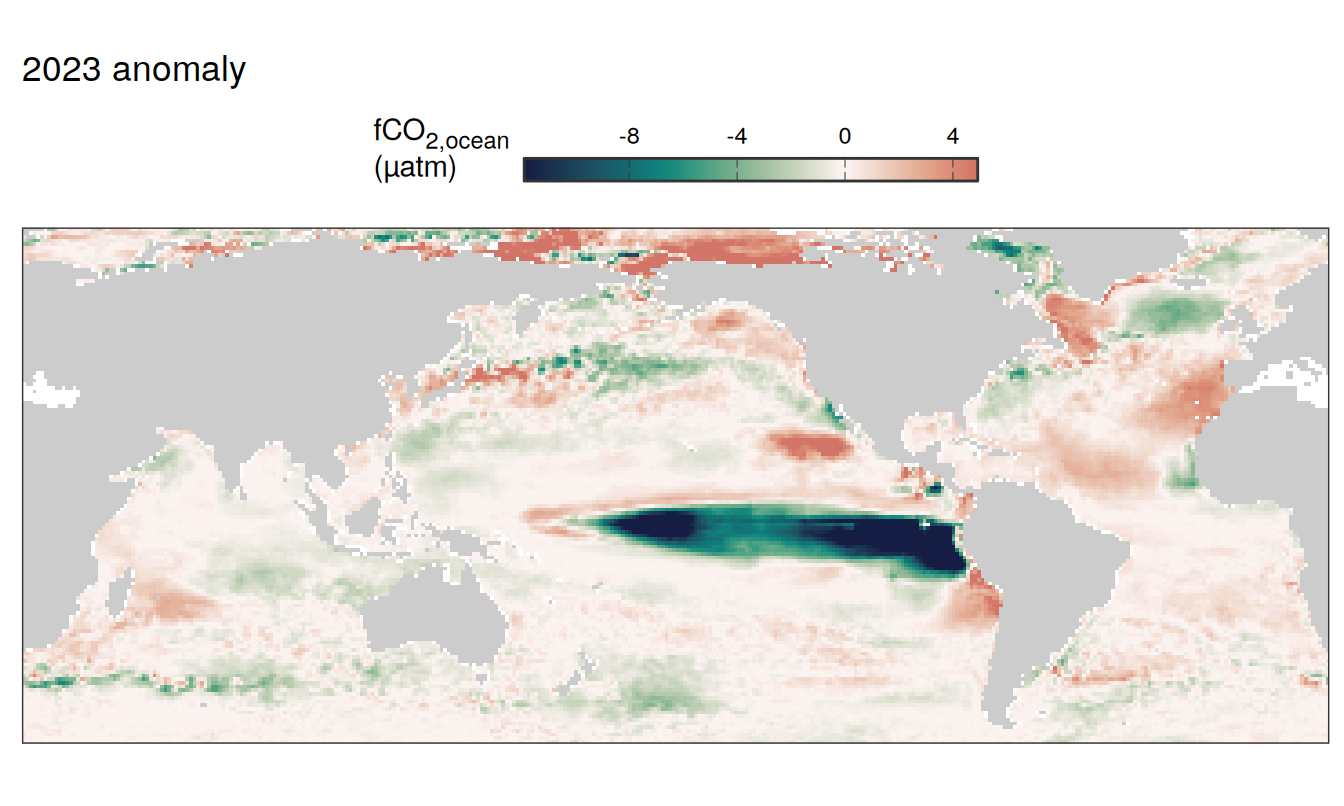
SST flux slope
pco2_product_map_annual_slope <-
pco2_product_map_annual_anomaly %>%
filter(year != 2023) %>%
select(year, lon, lat, resid, name) %>%
pivot_wider(values_from = resid) %>%
select(lon, lat, fgco2, temperature) %>%
drop_na() %>%
nest(data = -c(lon, lat)) %>%
mutate(fit = map(data, ~ flm(
formula = fgco2 ~ temperature, data = .x
)))
pco2_product_map_annual_slope <-
pco2_product_map_annual_slope %>%
unnest_wider(fit) %>%
select(lon, lat, slope = temperature) %>%
mutate(slope = as.vector(slope))
map +
geom_tile(data = pco2_product_map_annual_slope,
aes(lon, lat, fill = slope)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
limits = c(
quantile(pco2_product_map_annual_slope$slope,.01),
quantile(pco2_product_map_annual_slope$slope, .99)),
oob = squish
) +
labs(title = "Correlation of historic annual flux and SST anomalies") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")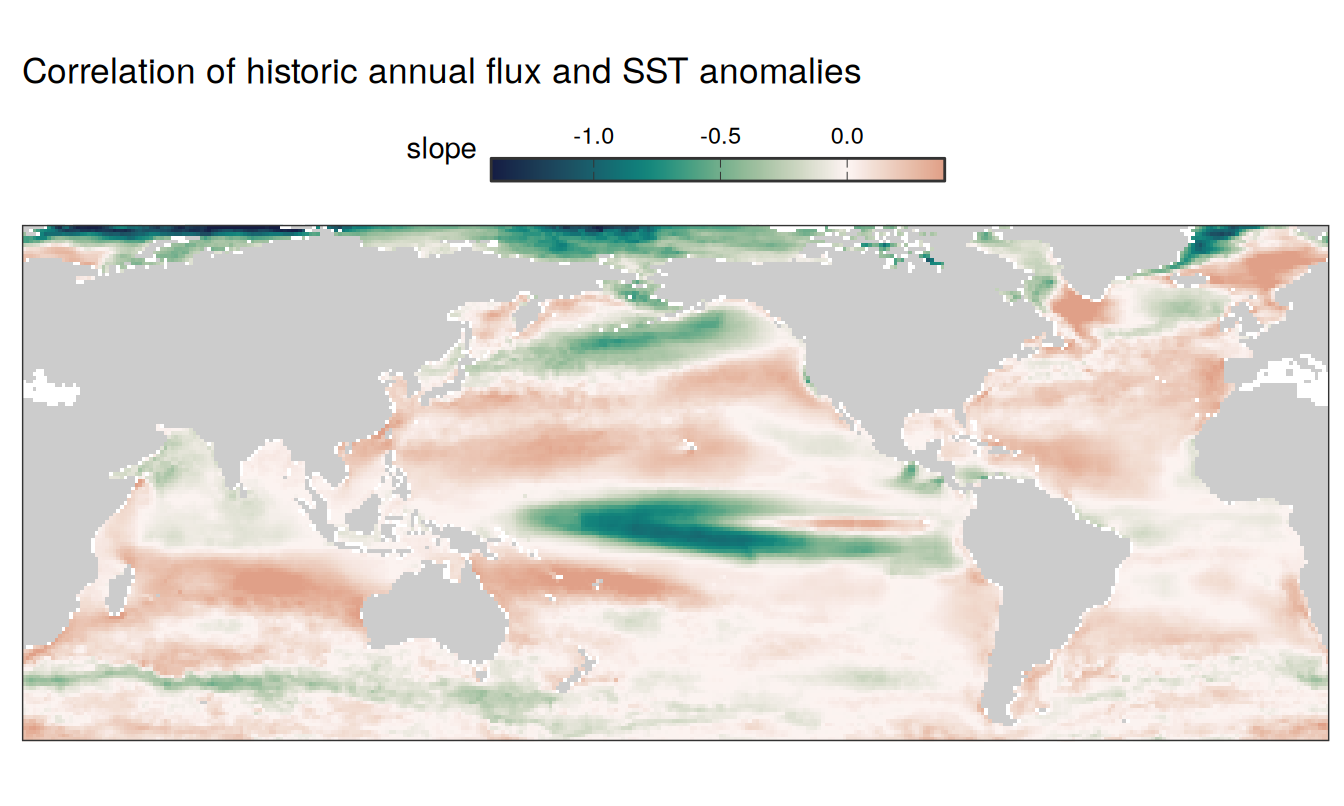
pco2_product_map_annual_anomaly_temperature_predict <-
inner_join(
pco2_product_map_annual_slope,
pco2_product_map_annual_anomaly %>%
filter(name %in% c("temperature", "fgco2")) %>%
select(lon, lat, name, resid, year) %>%
drop_na() %>%
arrange(desc(name)) %>%
pivot_wider(values_from = "resid")
)
pco2_product_map_annual_anomaly_temperature_predict <-
pco2_product_map_annual_anomaly_temperature_predict %>%
mutate(fgco2_predict = slope * temperature)
pco2_product_map_annual_anomaly_temperature_predict %>%
filter(year == 2023) %>%
select(-slope) %>%
pivot_longer(c(fgco2, temperature, fgco2_predict),
values_to = "resid") %>%
drop_na() %>%
# filter(name == "fgco2_predict") %>%
group_split(name) %>%
# head(1) %>%
map( ~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid)) +
labs(title = paste(2023,"anomaly")) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)[[1]]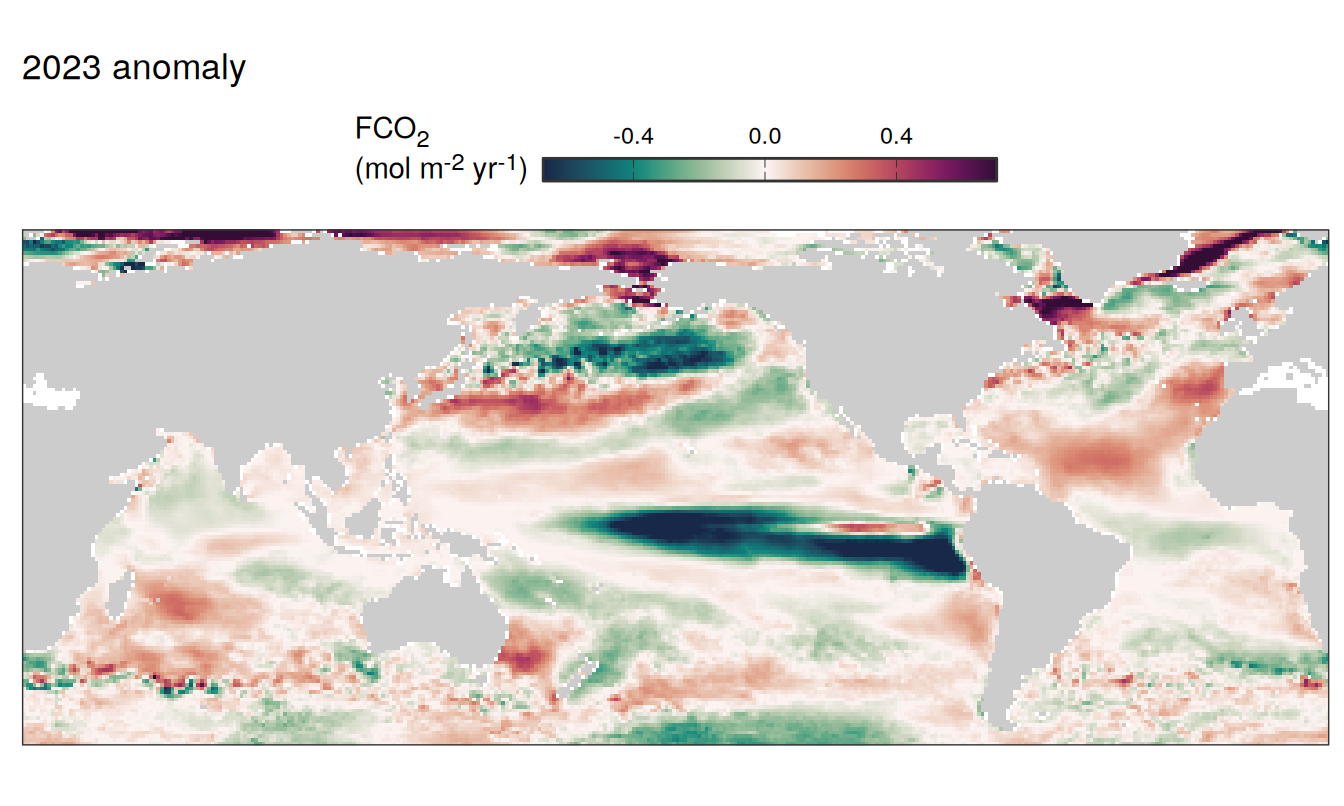
[[2]]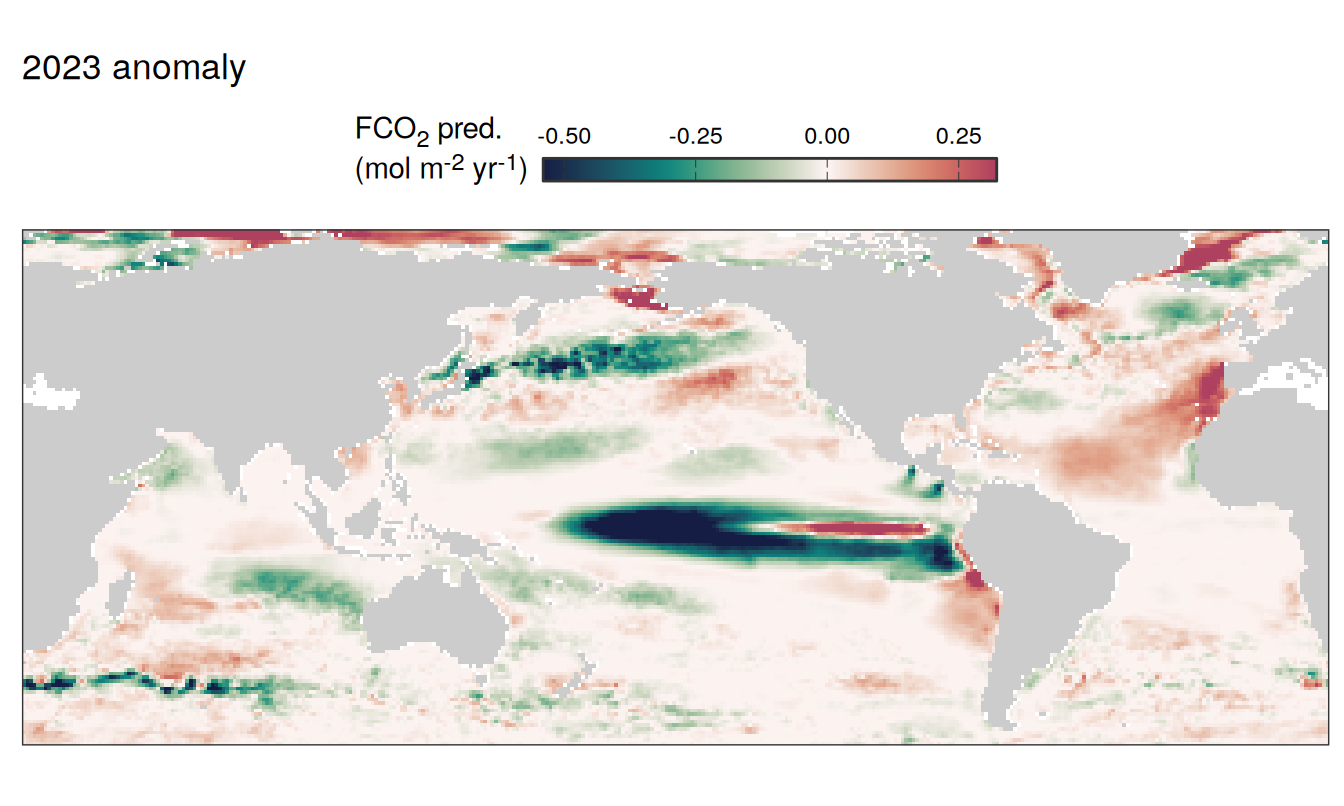
[[3]]
pco2_product_biome_annual_anomaly_temperature_predict <-
full_join(
pco2_product_map_annual_anomaly_temperature_predict %>%
select(lon, lat, year, temperature, fgco2, fgco2_predict),
biome_mask %>%
mutate(area = earth_surf(lat, lon))
)
pco2_product_biome_annual_anomaly_temperature_predict_global <-
pco2_product_biome_annual_anomaly_temperature_predict %>%
drop_na() %>%
mutate(fgco2_int = fgco2,
fgco2_predict_int = fgco2_predict) %>%
mutate(biome = case_when(str_detect(biome, "SO-SPSS|SO-ICE|Arctic") ~ "Polar",
TRUE ~ "Global non-polar")) %>%
filter(biome == "Global non-polar") %>%
select(-c(lon, lat)) %>%
group_by(year, biome) %>%
summarise(across(-c(fgco2_int, fgco2_predict_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(c(fgco2_int, fgco2_predict_int),
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_annual_anomaly_temperature_predict <-
pco2_product_biome_annual_anomaly_temperature_predict %>%
drop_na() %>%
mutate(fgco2_int = fgco2,
fgco2_predict_int = fgco2_predict) %>%
select(-c(lon, lat)) %>%
group_by(year, biome) %>%
summarise(across(-c(fgco2_int, fgco2_predict_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(c(fgco2_int, fgco2_predict_int),
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_annual_anomaly_temperature_predict <-
bind_rows(pco2_product_biome_annual_anomaly_temperature_predict_global,
pco2_product_biome_annual_anomaly_temperature_predict)
rm(pco2_product_biome_annual_anomaly_temperature_predict_global)
pco2_product_biome_annual_anomaly_temperature_predict <-
pco2_product_biome_annual_anomaly_temperature_predict %>%
filter(!is.na(biome))
pco2_product_biome_annual_anomaly_temperature_predict %>%
select(year, biome, fgco2_int, fgco2_predict_int) %>%
filter(biome == "Global non-polar") %>%
pivot_longer(starts_with("fgco2"),
values_to = "resid") %>%
ggplot(aes(year, resid, col = name)) +
geom_path() +
geom_point()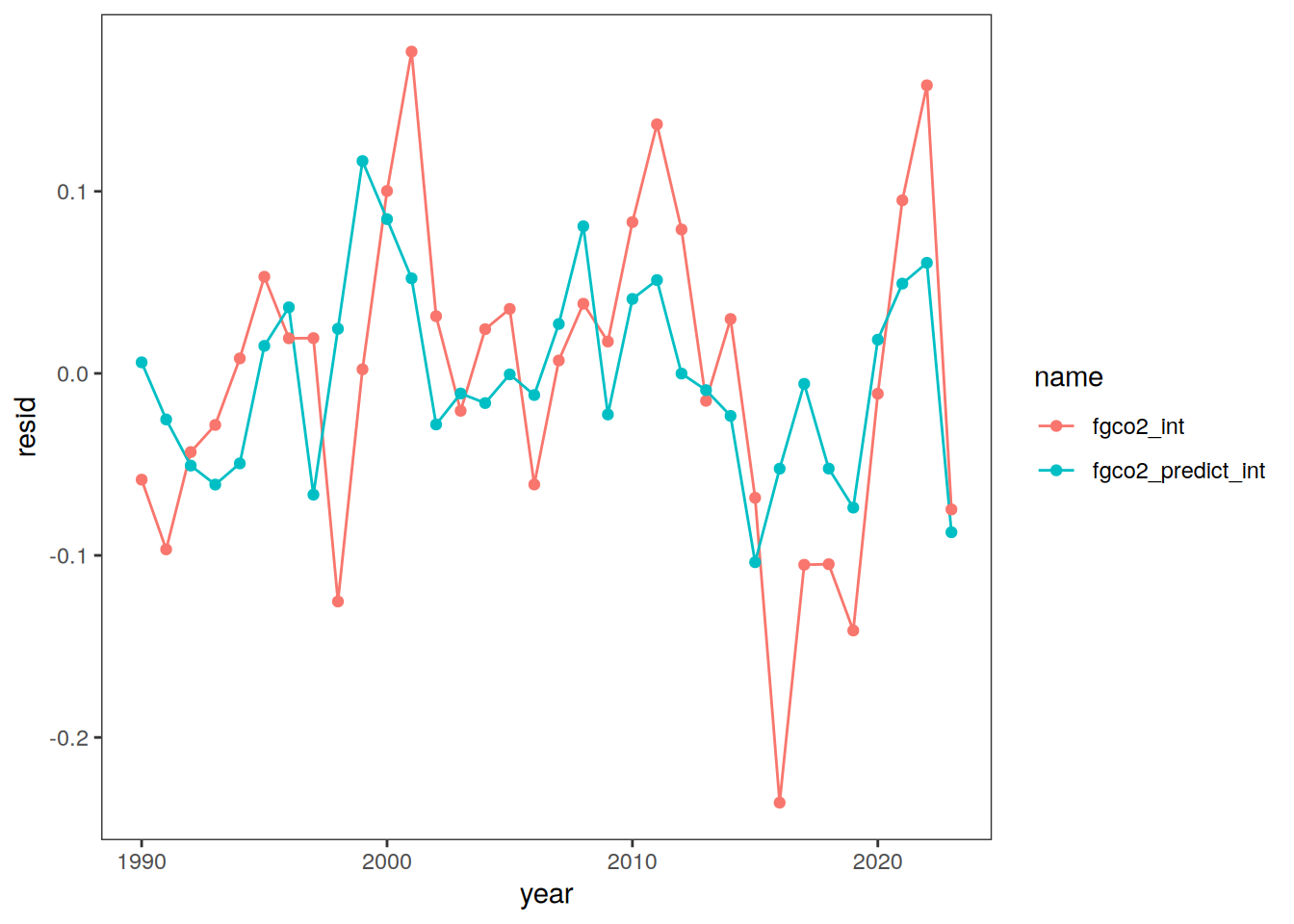
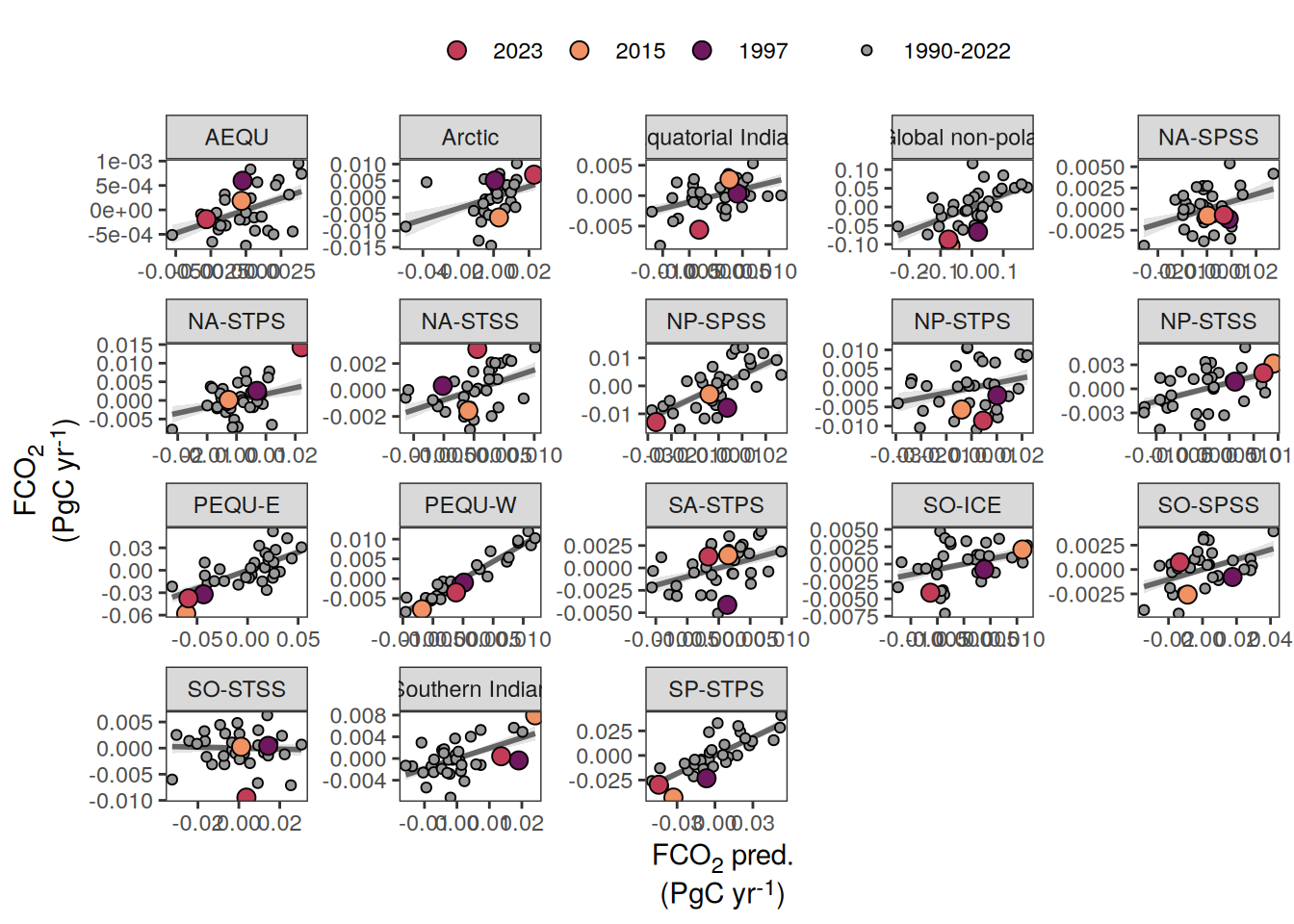
pco2_product_biome_annual_anomaly_temperature_predict %>%
ggplot(aes(fgco2_int, fgco2_predict_int)) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_point(data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(fill = "1990-2022"),
shape = 21) +
scale_color_manual(values = "grey60", name = "X") +
scale_fill_manual(values = "grey60", name = "X") +
new_scale_fill() +
new_scale_color() +
geom_point(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(fill = as.factor(year)),
shape = 21,
size = 3
) +
scale_fill_manual(
values = rev(warm_cool_gradient[c(17,13,20)]),
guide = guide_legend(reverse = TRUE,
order = 2)
) +
scale_color_manual(
values = rev(warm_cool_gradient[c(17,13,20)]),
guide = guide_legend(reverse = TRUE,
order = 2)
) +
labs(y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks("fgco2_predict_int")$i_legend_title) +
facet_wrap(~ biome, scales = "free") +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)
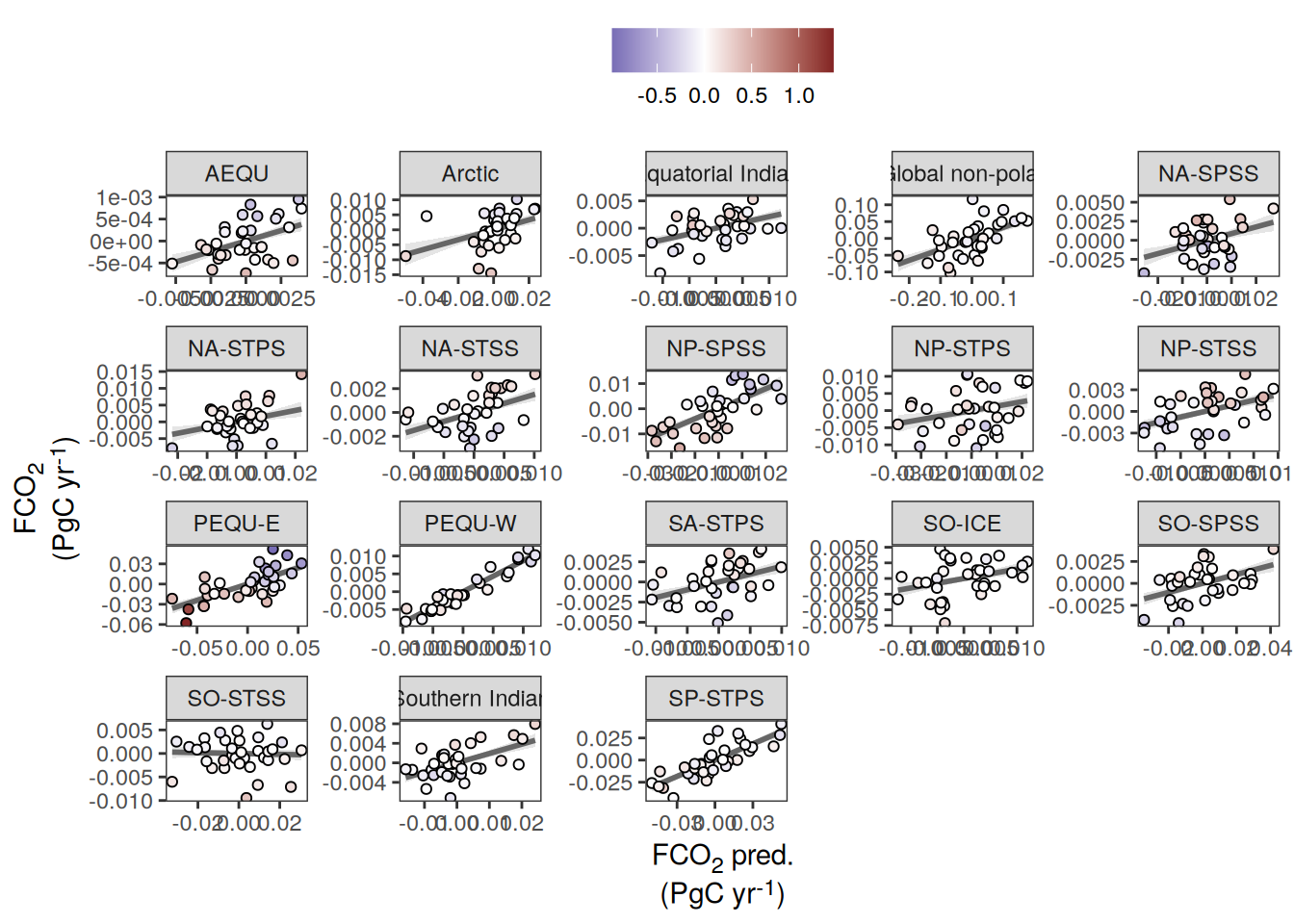
pco2_product_biome_annual_anomaly_temperature_predict %>%
ggplot(aes(fgco2_int, fgco2_predict_int)) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_point(aes(fill = temperature),
shape = 21) +
scale_fill_divergent() +
labs(y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks("fgco2_predict_int")$i_legend_title) +
facet_wrap(~ biome, scales = "free") +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)
pco2_product_map_annual_anomaly %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_map_annual_anomaly.csv"
)
)
pco2_product_map_annual_anomaly_temperature_predict %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_map_annual_anomaly_temperature_predict.csv"
)
)
pco2_product_biome_annual_anomaly_temperature_predict %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_annual_anomaly_temperature_predict.csv"
)
)
rm(pco2_product_map_annual_anomaly,
pco2_product_map_annual_slope,
pco2_product_map_annual_anomaly_temperature_predict,
pco2_product_biome_annual_anomaly_temperature_predict)
gc()Monthly means
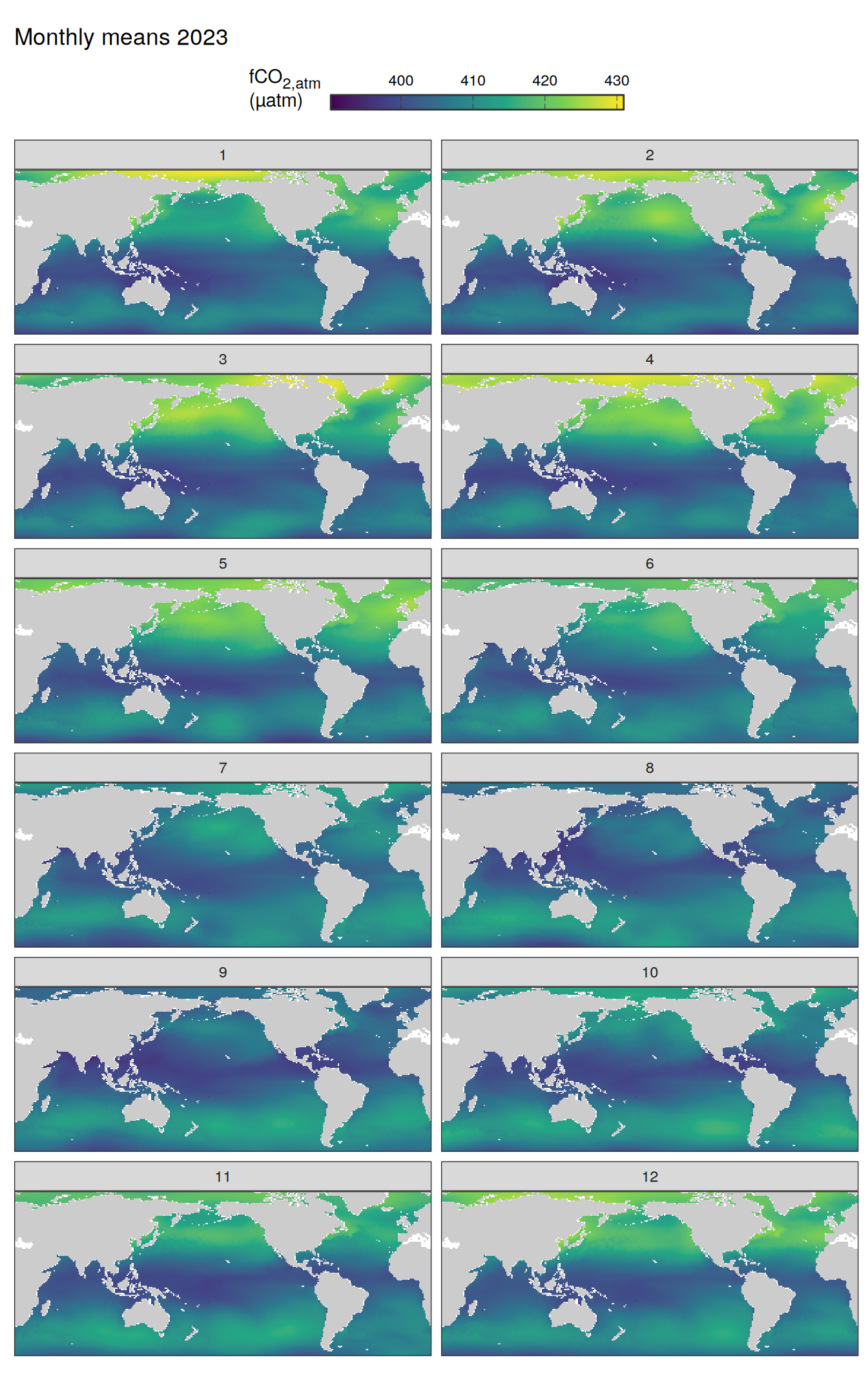
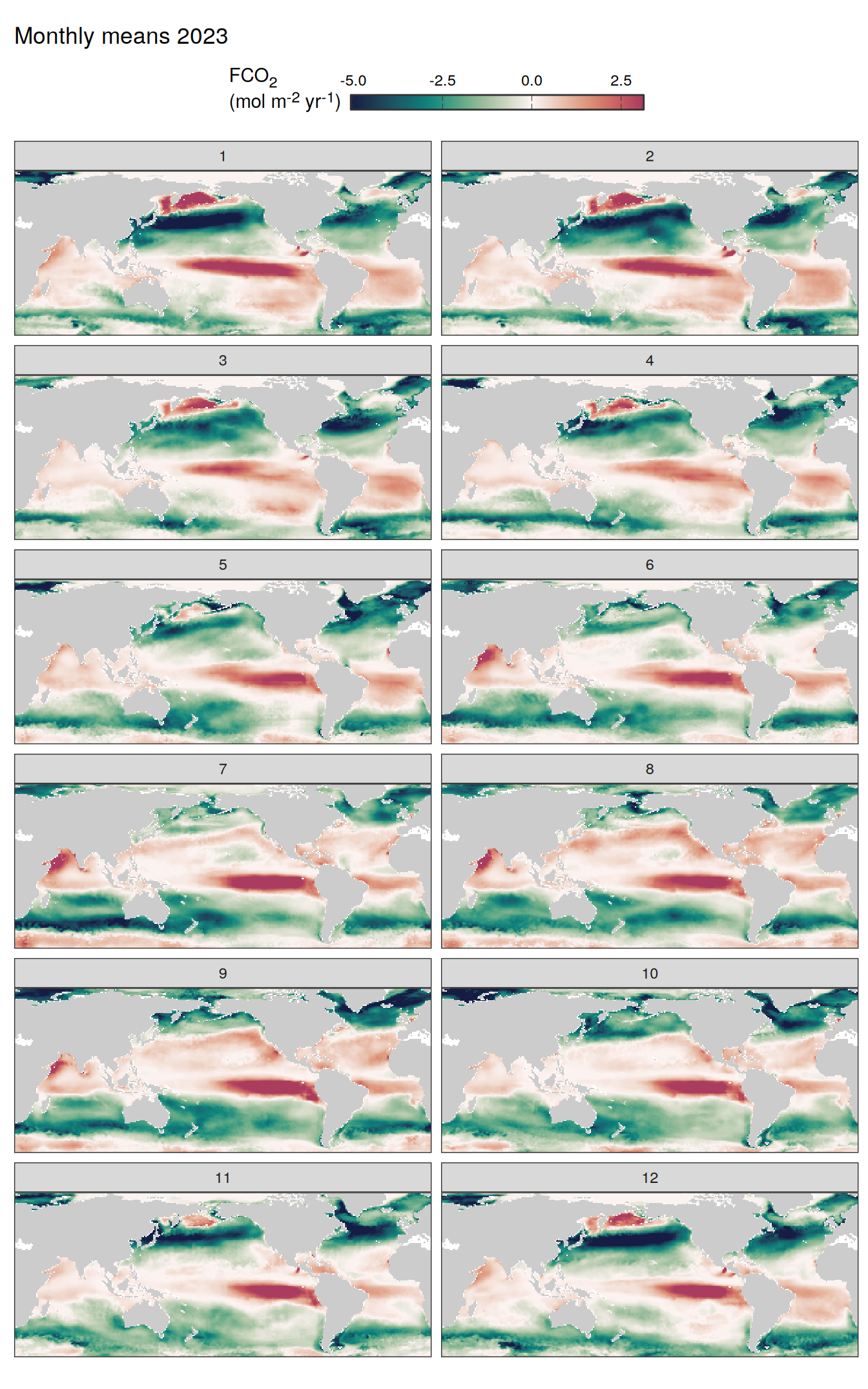
2023 absolute
pco2_product_map_monthly_anomaly <-
pco2_product_map_monthly %>%
drop_na() %>%
anomaly_determination(lon, lat, month)
pco2_product_map_monthly_anomaly <-
pco2_product_map_monthly_anomaly %>%
drop_na()
pco2_product_map_monthly_anomaly %>%
filter(year == 2023, !(name %in% name_divergent)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = value)) +
labs(title = paste("Monthly means", 2023)) +
scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap(~ month, ncol = 2)
)
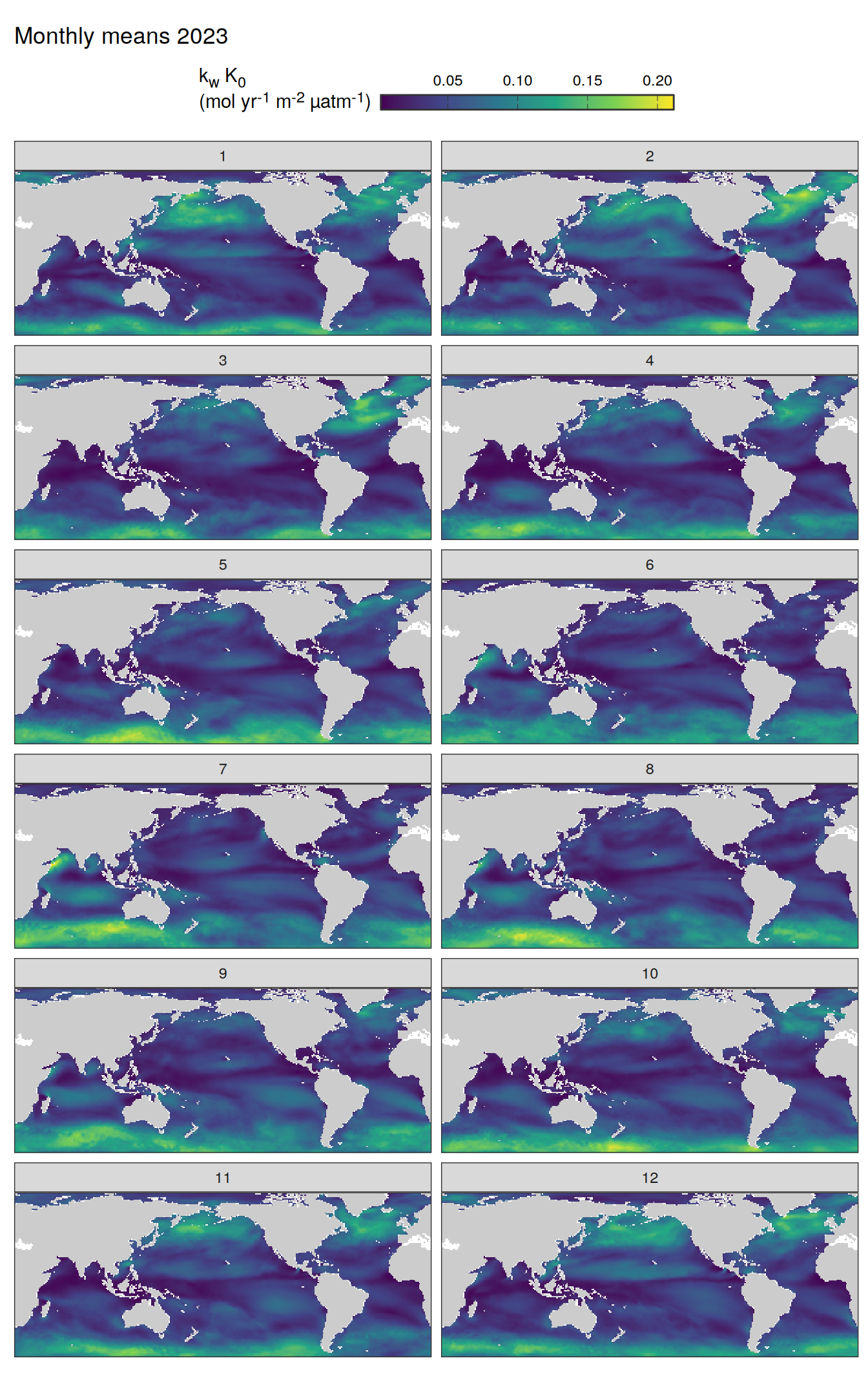
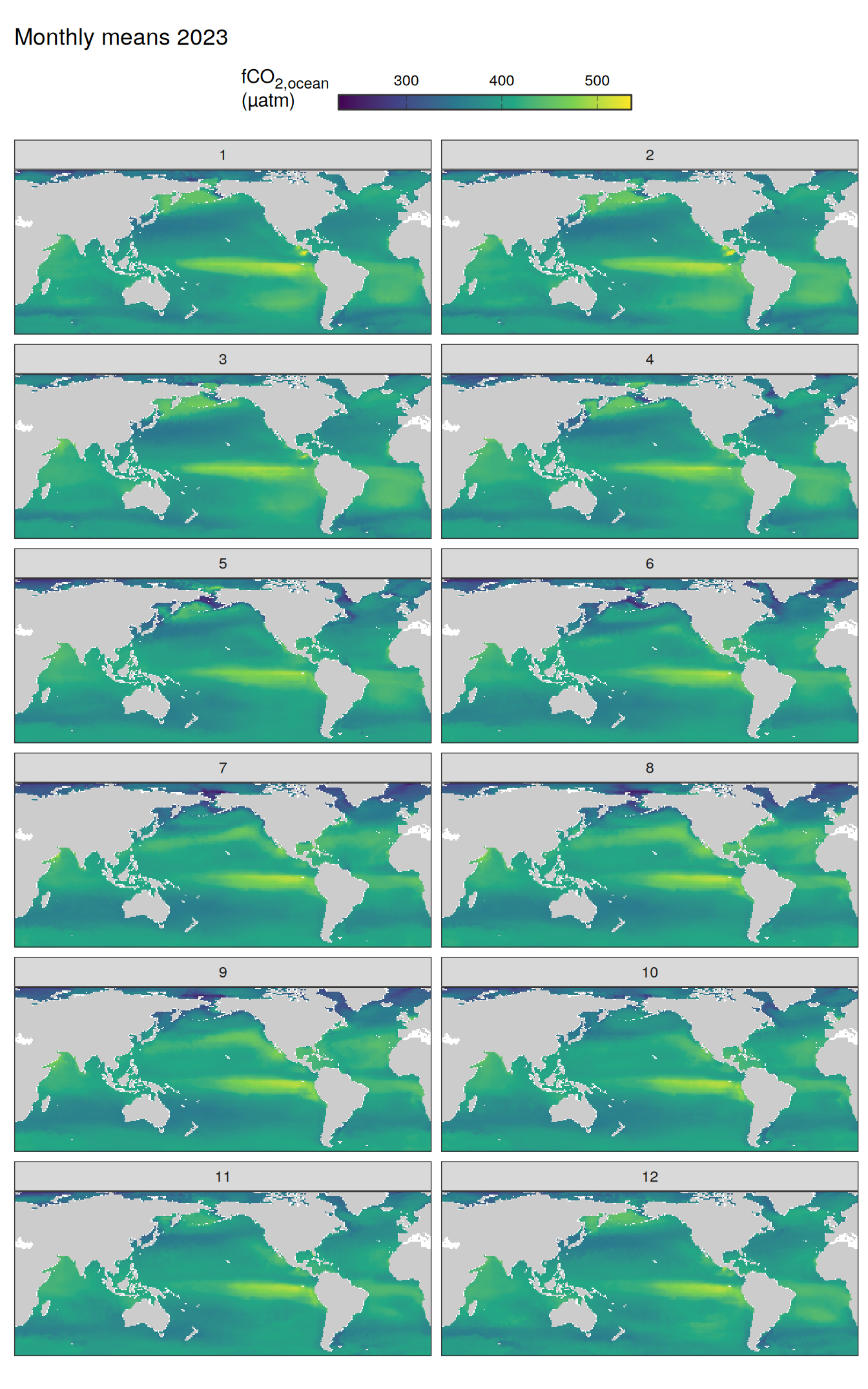
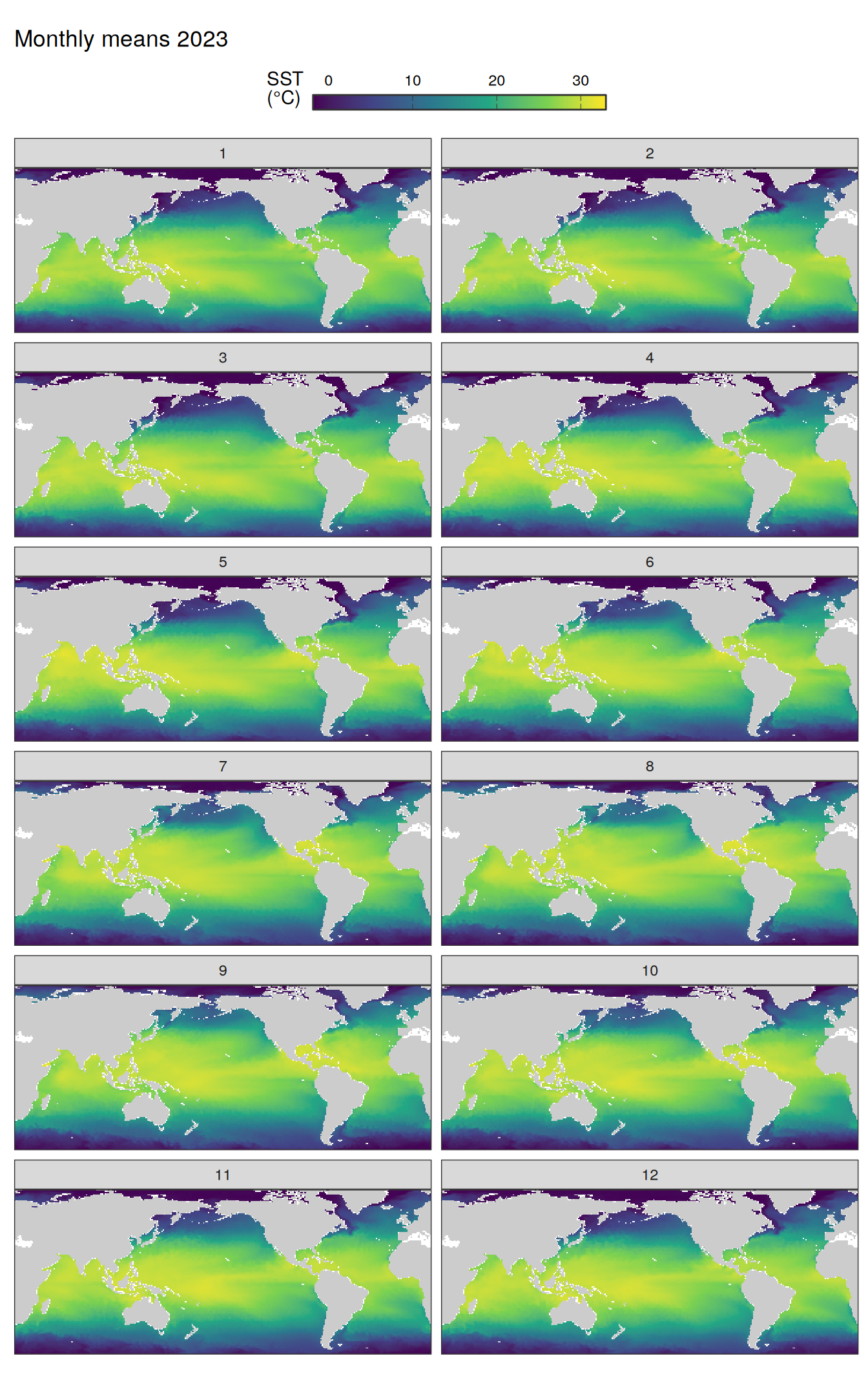
pco2_product_map_monthly_anomaly %>%
filter(year == 2023, name %in% name_divergent) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = value)) +
labs(title = paste("Monthly means", 2023)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap( ~ month, ncol = 2)
)
Trends
pco2_product_map_monthly_anomaly %>%
group_by(name) %>%
filter(year %in% c(min(year), max(year))) %>%
ungroup() %>%
select(-c(value, resid)) %>%
arrange(year) %>%
group_by(lon, lat, name, month) %>%
mutate(change = fit - lag(fit),
period = paste(lag(year), year, sep = "-")) %>%
ungroup() %>%
filter(!is.na(change)) %>%
group_split(name) %>%
head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = change)) +
labs(title = paste("Change: ", .x$period)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$change, .01), quantile(.x$change, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap(~ month, ncol = 2)
)2023 anomaly
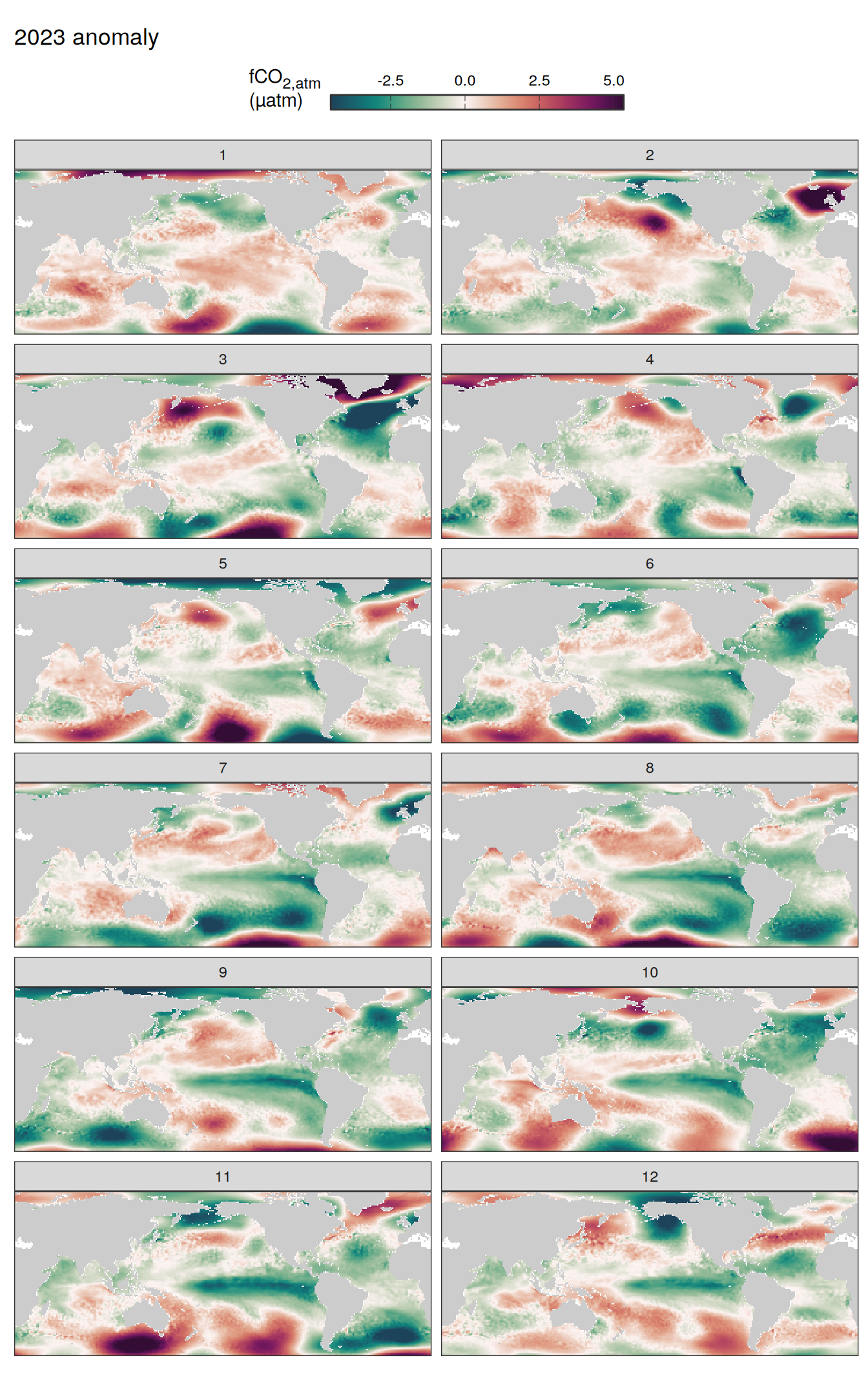
pco2_product_map_monthly_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = resid)) +
labs(title = paste(2023, "anomaly")) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap( ~ month, ncol = 2)
)
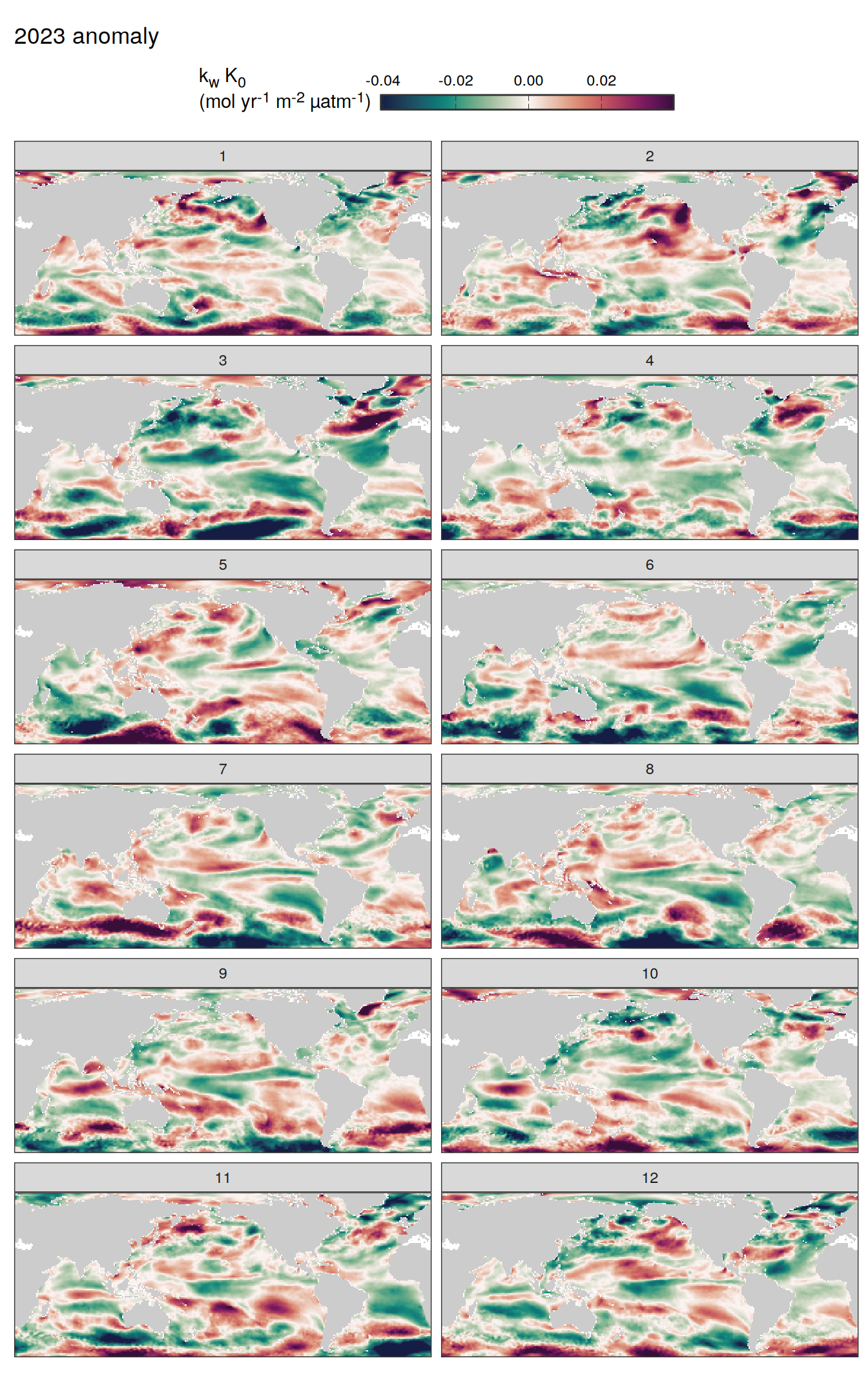
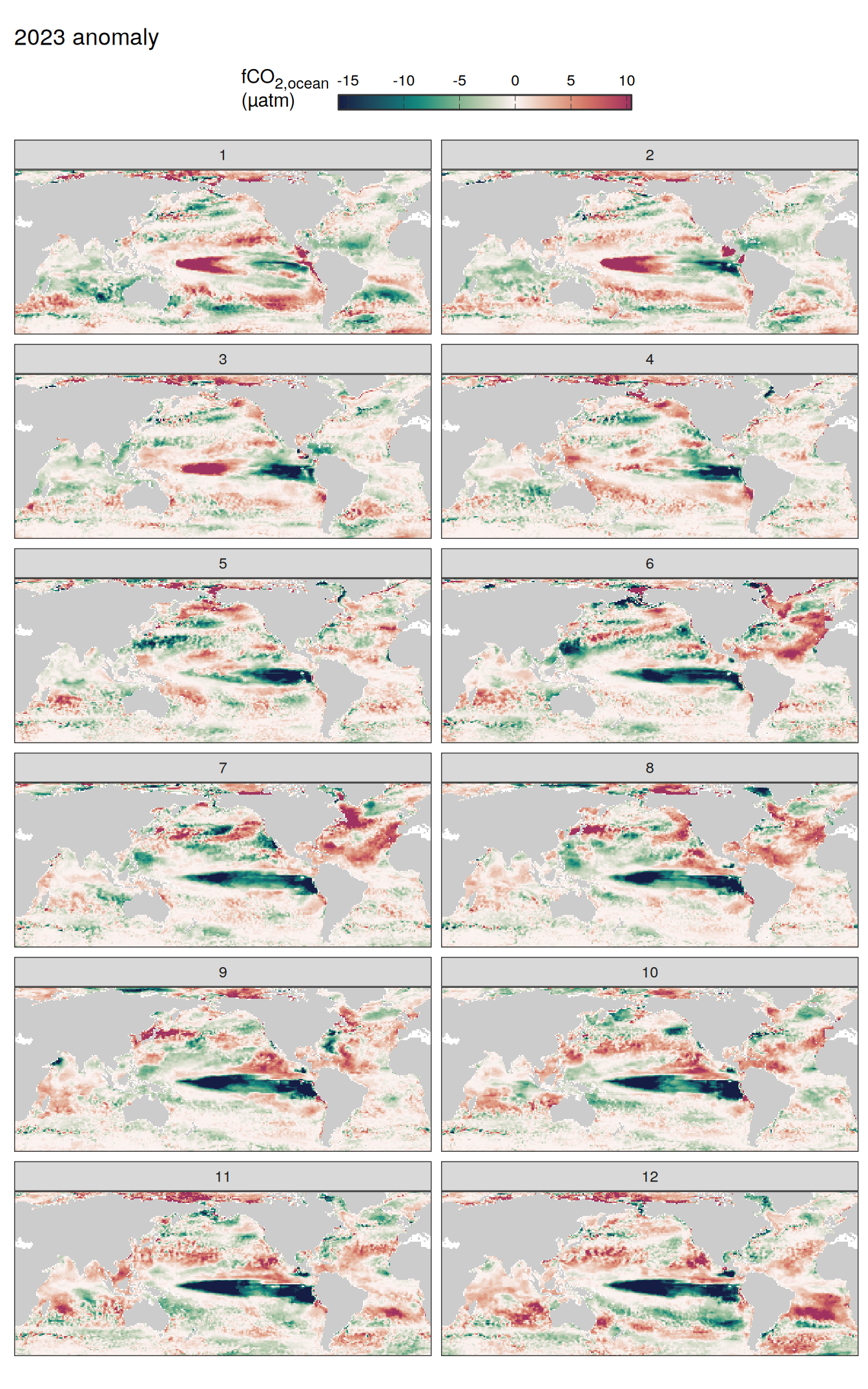
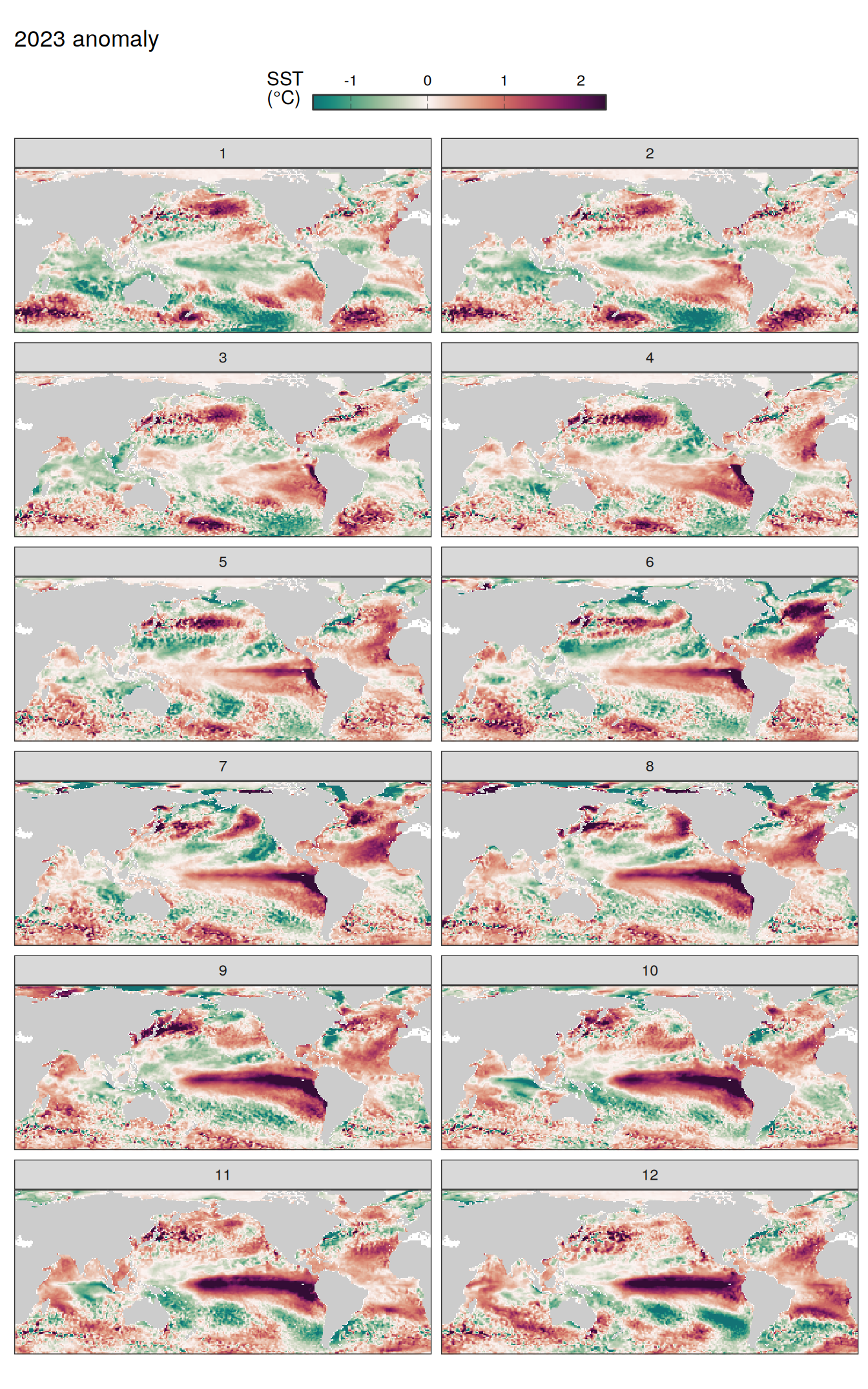
fCO2 decomposition
pco2_product_map_monthly_fCO2_decomposition <-
fco2_decomposition(pco2_product_map_monthly_anomaly,
year, month, lon, lat)
# pco2_product_map_monthly_fCO2_decomposition %>%
# filter(year == 2023) %>%
# mutate(product == "pco2 product") %>%
# group_split(product) %>%
# head(1) %>%
# map(
# ~ map +
# geom_tile(data = .x,
# aes(lon, lat, fill = resid)) +
# labs(title = .x$product) +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks("sfco2"),
# limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
# oob = squish
# ) +
# facet_grid(month ~ name,
# labeller = labeller(name = x_axis_labels)) +
# guides(
# fill = guide_colorbar(
# barheight = unit(0.3, "cm"),
# barwidth = unit(6, "cm"),
# ticks = TRUE,
# ticks.colour = "grey20",
# frame.colour = "grey20",
# label.position = "top",
# direction = "horizontal"
# )
# ) +
# theme(legend.title = element_markdown(),
# legend.position = "top")
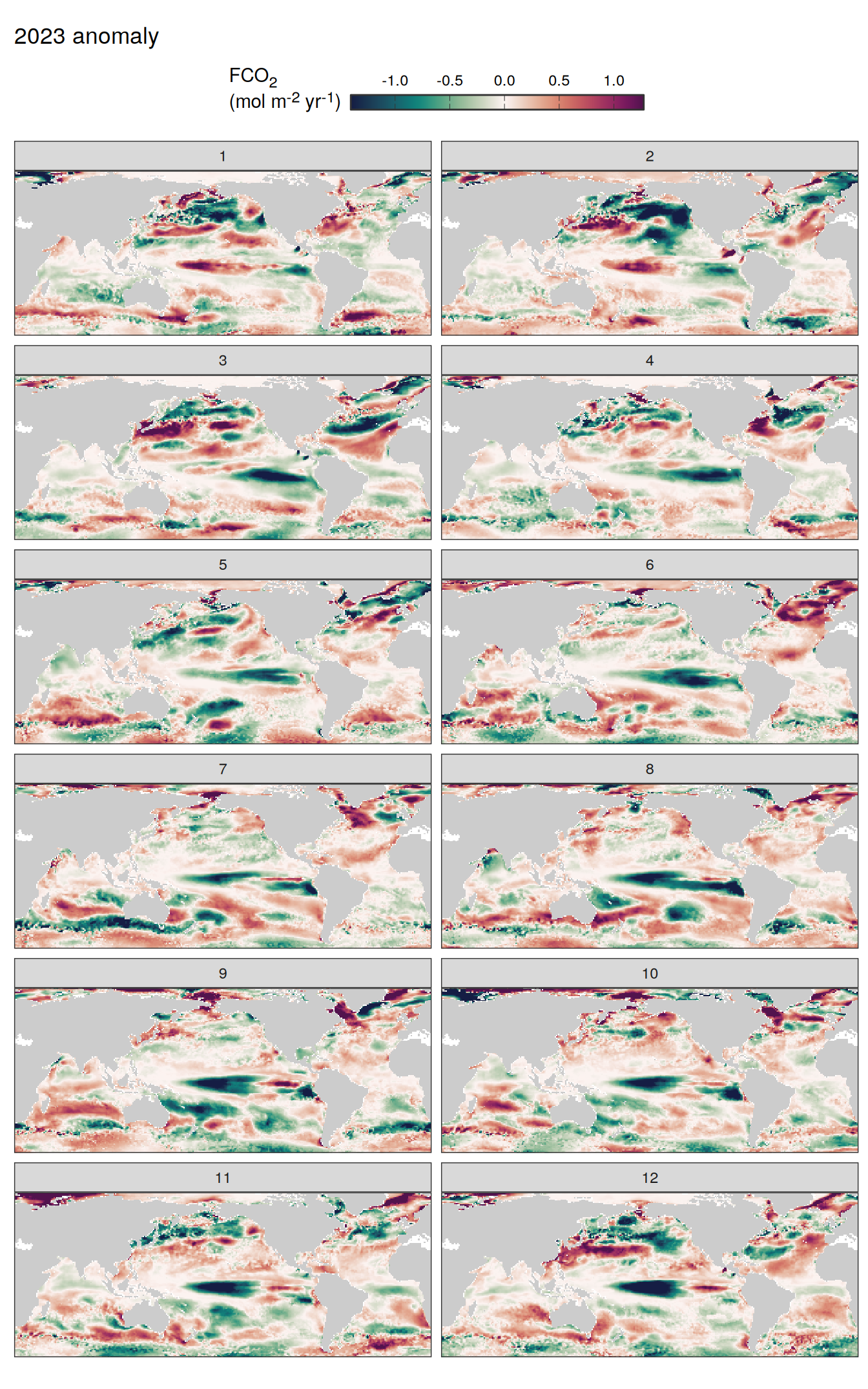
# )pco2_product_map_annual_fCO2_decomposition <-
pco2_product_map_monthly_fCO2_decomposition %>%
select(year, lat, lon, name, resid) %>%
fgroup_by(year, lat, lon, name) %>%
fmean()
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4291288 229.2 27299156 1458.0 104138011 5561.6
Vcells 1683142661 12841.4 3844364568 29330.2 3844324302 29329.9map +
geom_tile(data = pco2_product_map_annual_fCO2_decomposition %>%
filter(year == 2023), aes(lon, lat, fill = resid)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("sfco2"),
limits = c(
quantile(pco2_product_map_annual_fCO2_decomposition$resid, .01),
quantile(pco2_product_map_annual_fCO2_decomposition$resid, .99)
),
oob = squish
) +
facet_wrap(~ name,
ncol = 2,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")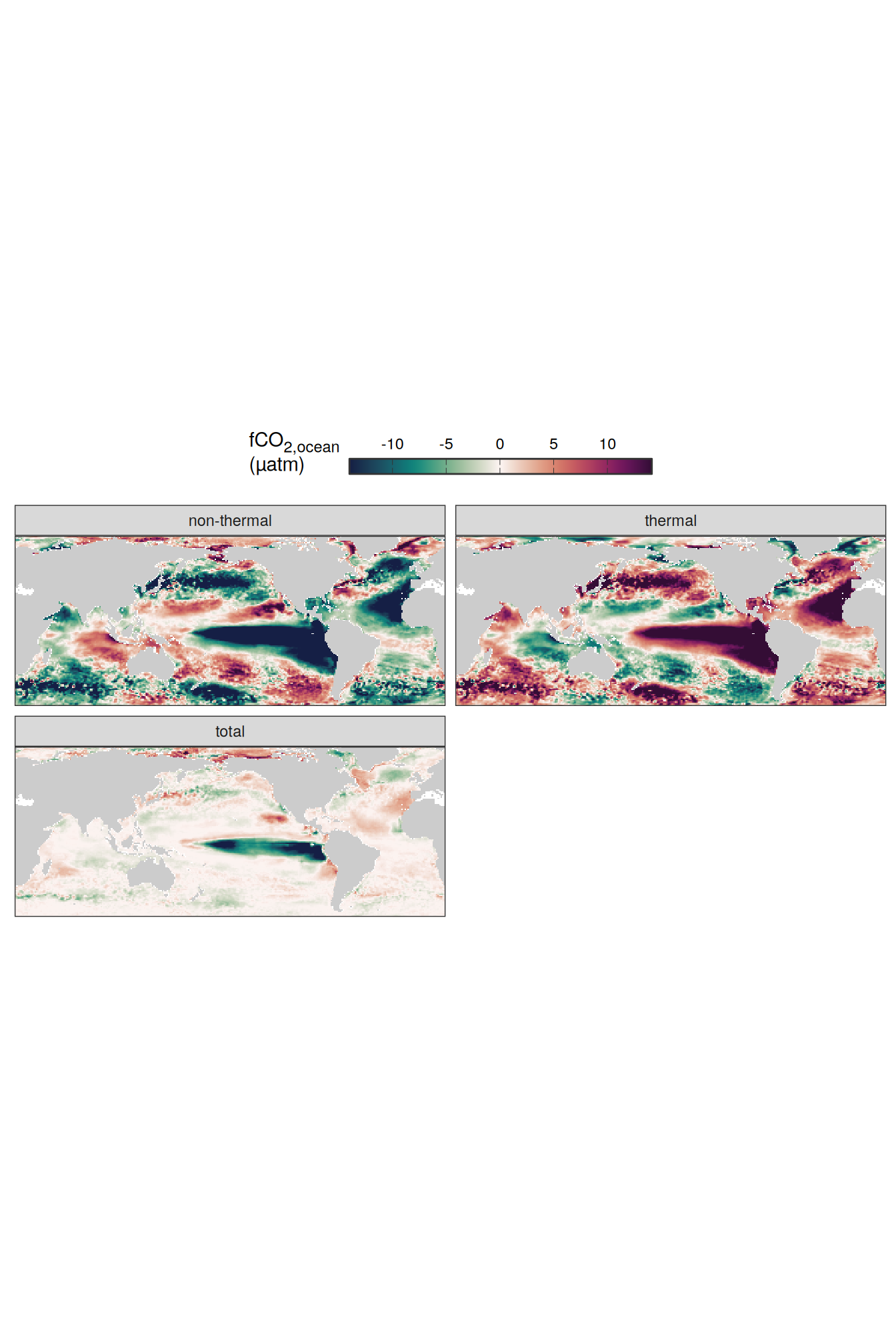
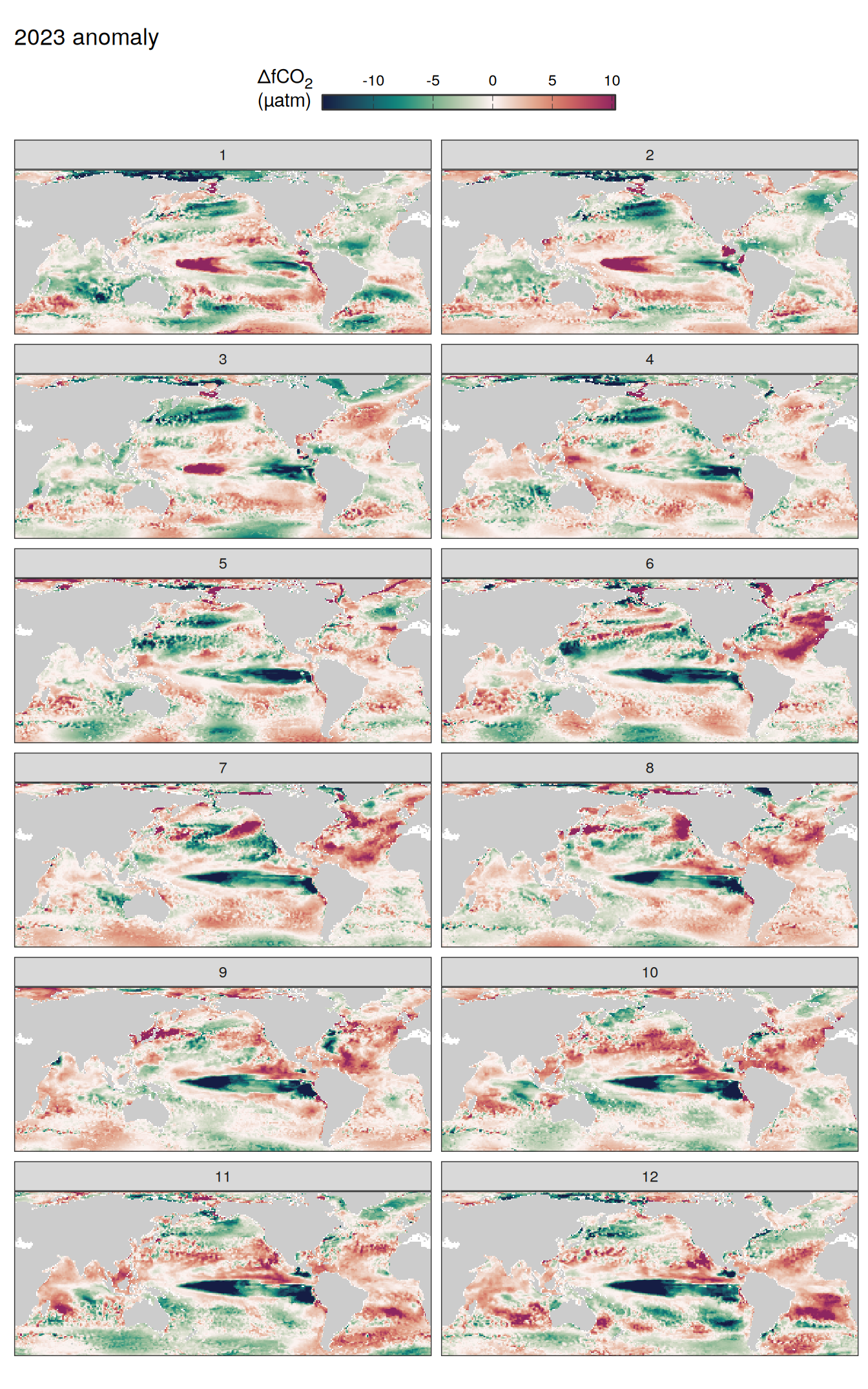
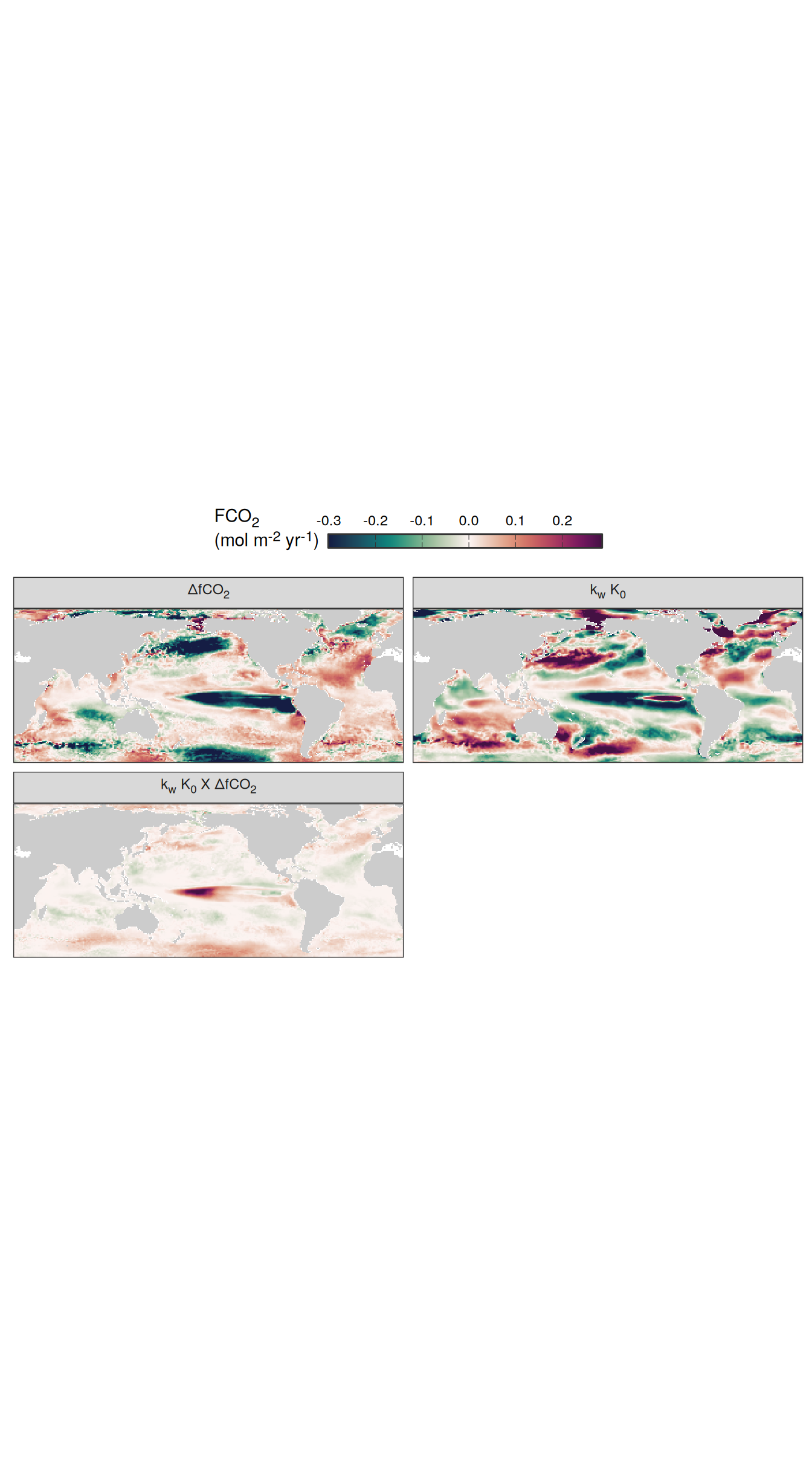
Flux attribution
pco2_product_map_monthly_flux_attribution <-
flux_attribution(pco2_product_map_monthly_anomaly,
year, month, lon, lat)
# pco2_product_map_monthly_flux_attribution %>%
# filter(year == 2023) %>%
# drop_na() %>%
# mutate(product == "pco2 product") %>%
# group_split(product) %>%
# head(1) %>%
# map(
# ~ map +
# geom_tile(data = .x,
# aes(lon, lat, fill = resid)) +
# labs(subtitle = .x$product) +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks("fgco2"),
# limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
# oob = squish
# ) +
# theme(legend.title = element_markdown(),
# legend.position = "bottom") +
# facet_grid(month ~ name,
# labeller = labeller(name = x_axis_labels)) +
# guides(
# fill = guide_colorbar(
# barheight = unit(0.3, "cm"),
# barwidth = unit(6, "cm"),
# ticks = TRUE,
# ticks.colour = "grey20",
# frame.colour = "grey20",
# label.position = "top",
# direction = "horizontal"
# )
# ) +
# theme(legend.title = element_markdown(),
# legend.position = "top",
# strip.text.x.top = element_markdown())
# )pco2_product_map_annual_flux_attribution <-
pco2_product_map_monthly_flux_attribution %>%
group_by(year, lat, lon, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup()
map +
geom_tile(data = pco2_product_map_annual_flux_attribution %>%
filter(year == 2023), aes(lon, lat, fill = resid)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("fgco2"),
limits = c(
quantile(pco2_product_map_annual_flux_attribution$resid, .01, na.rm = TRUE),
quantile(pco2_product_map_annual_flux_attribution$resid, .99, na.rm = TRUE)
),
oob = squish
) +
theme(legend.title = element_markdown(), legend.position = "bottom") +
facet_wrap(~ name,
ncol = 2,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.title = element_markdown(),
legend.position = "top",
strip.text.x.top = element_markdown()
)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4309498 230.2 21839325 1166.4 104138011 5561.6
Vcells 1938693021 14791.1 3844364568 29330.2 3844324302 29329.9pco2_product_map_monthly_anomaly %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_map_monthly_anomaly.csv"
)
)
pco2_product_map_annual_flux_attribution %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_map_annual_flux_attribution.csv"
)
)
pco2_product_map_annual_fCO2_decomposition %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_map_annual_fCO2_decomposition.csv"
)
)
pco2_product_map_monthly_flux_attribution %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_map_monthly_flux_attribution.csv"
)
)
pco2_product_map_monthly_fCO2_decomposition %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_map_monthly_fCO2_decomposition.csv"
)
)
rm(pco2_product_map_annual_flux_attribution,
pco2_product_map_annual_fCO2_decomposition)
gc()Hovmoeller plots
The following Hovmoeller plots show the anomalies from the prediction of the linear/quadratic fits.
Hovmoeller plots are first presented as annual means, and than as monthly means. Note that the predictions for the monthly Hovmoeller plots are done individually for each month, such the mean seasonal anomaly from the annual mean is removed.
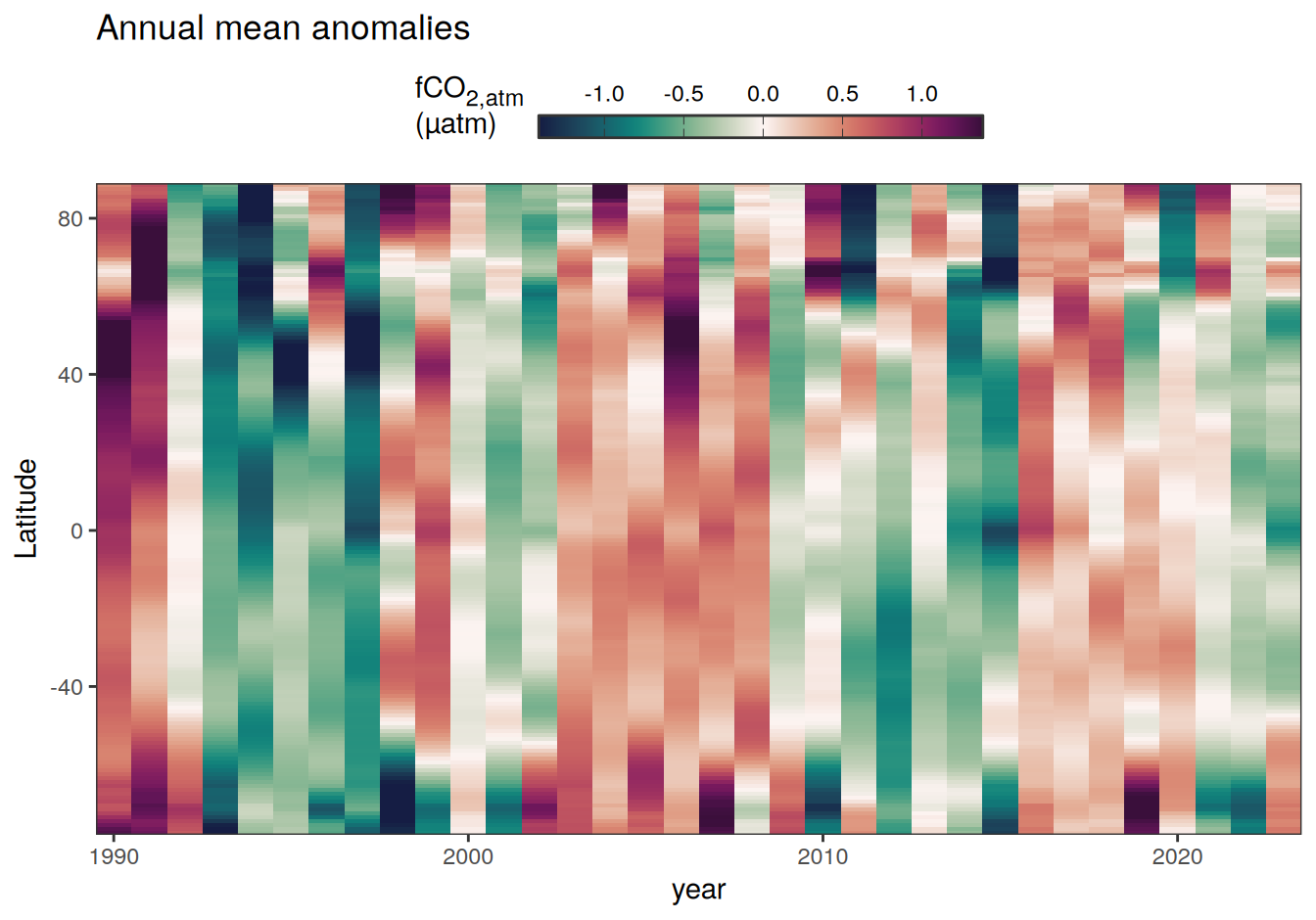
2023 annual anomalies
pco2_product_hovmoeller_annual_anomaly <-
pco2_product_hovmoeller_annual %>%
anomaly_determination(lat) %>%
filter(!is.na(resid))
pco2_product_hovmoeller_annual_anomaly %>%
# filter(name == "mld") %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x, aes(year, lat, fill = resid)) +
geom_raster() +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
coord_cartesian(expand = 0) +
labs(title = "Annual mean anomalies", y = "Latitude") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")
)
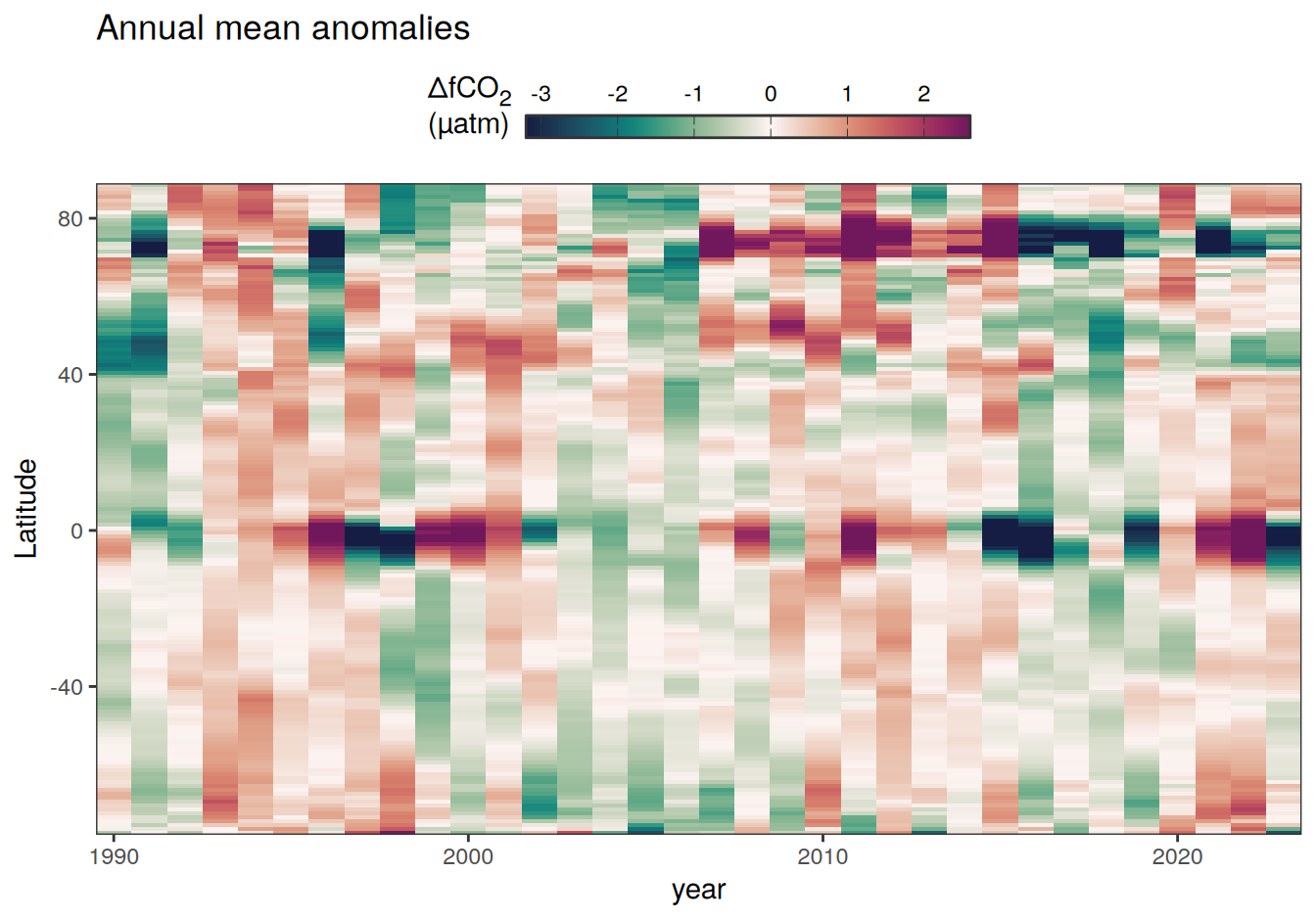
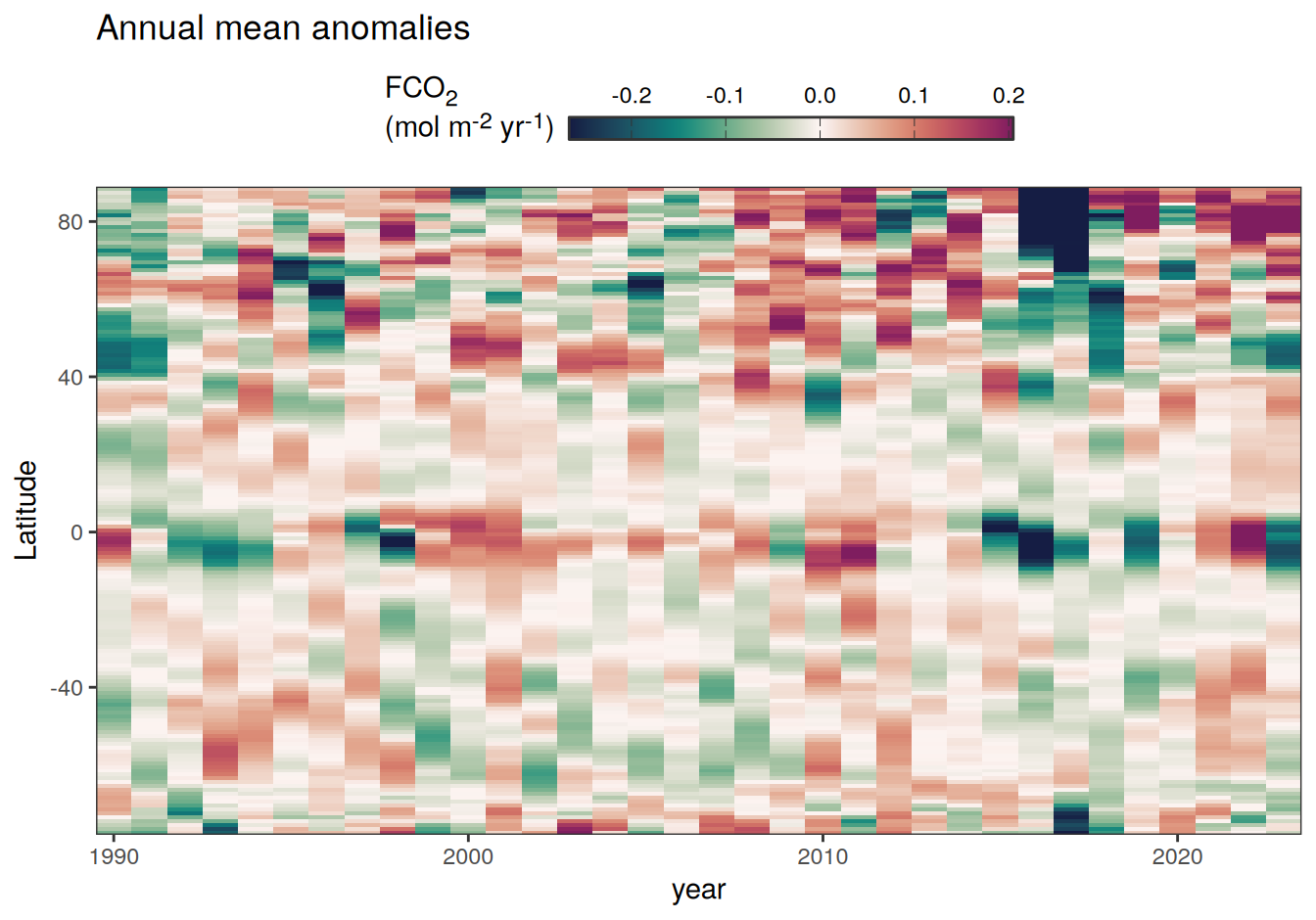
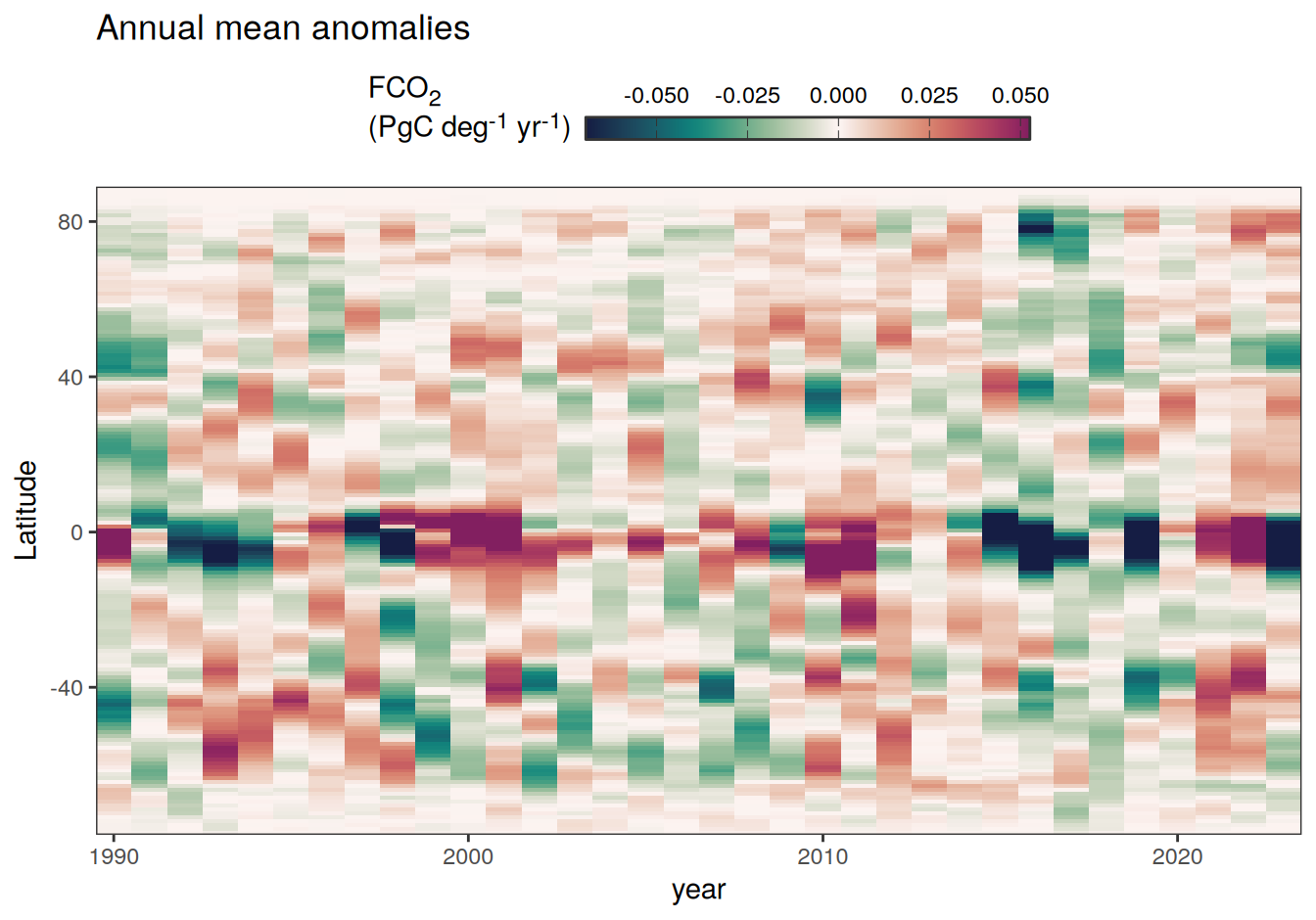
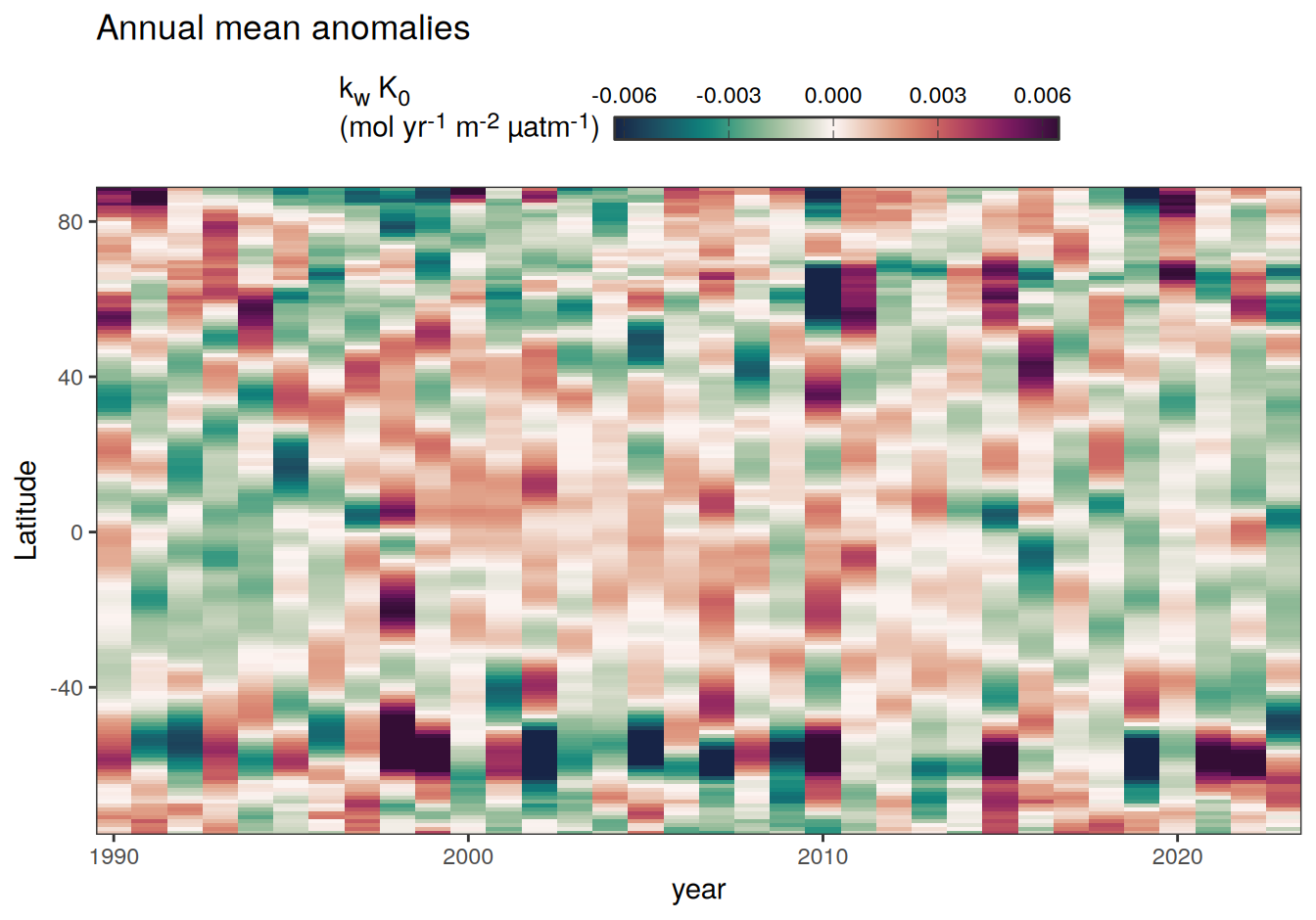
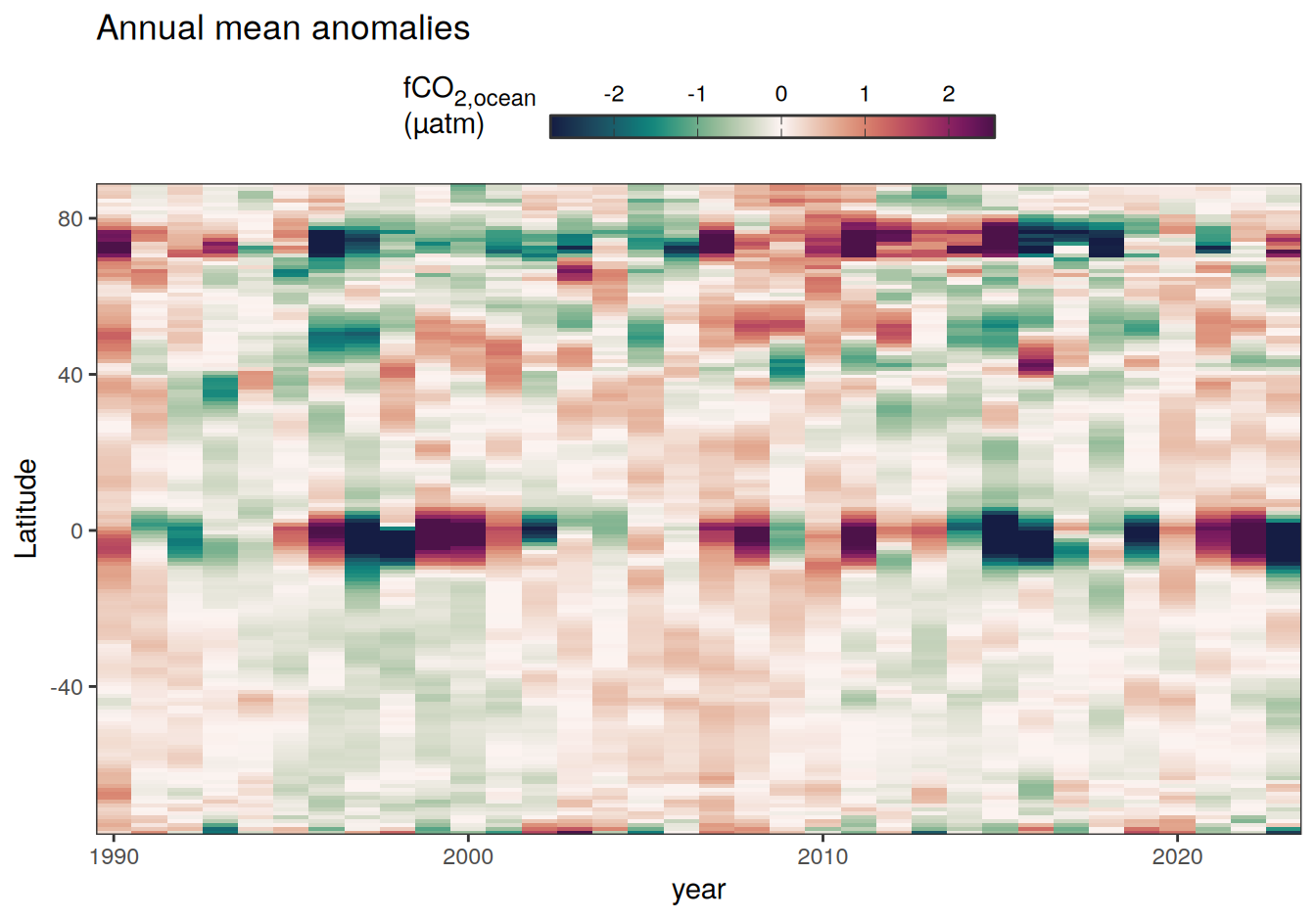
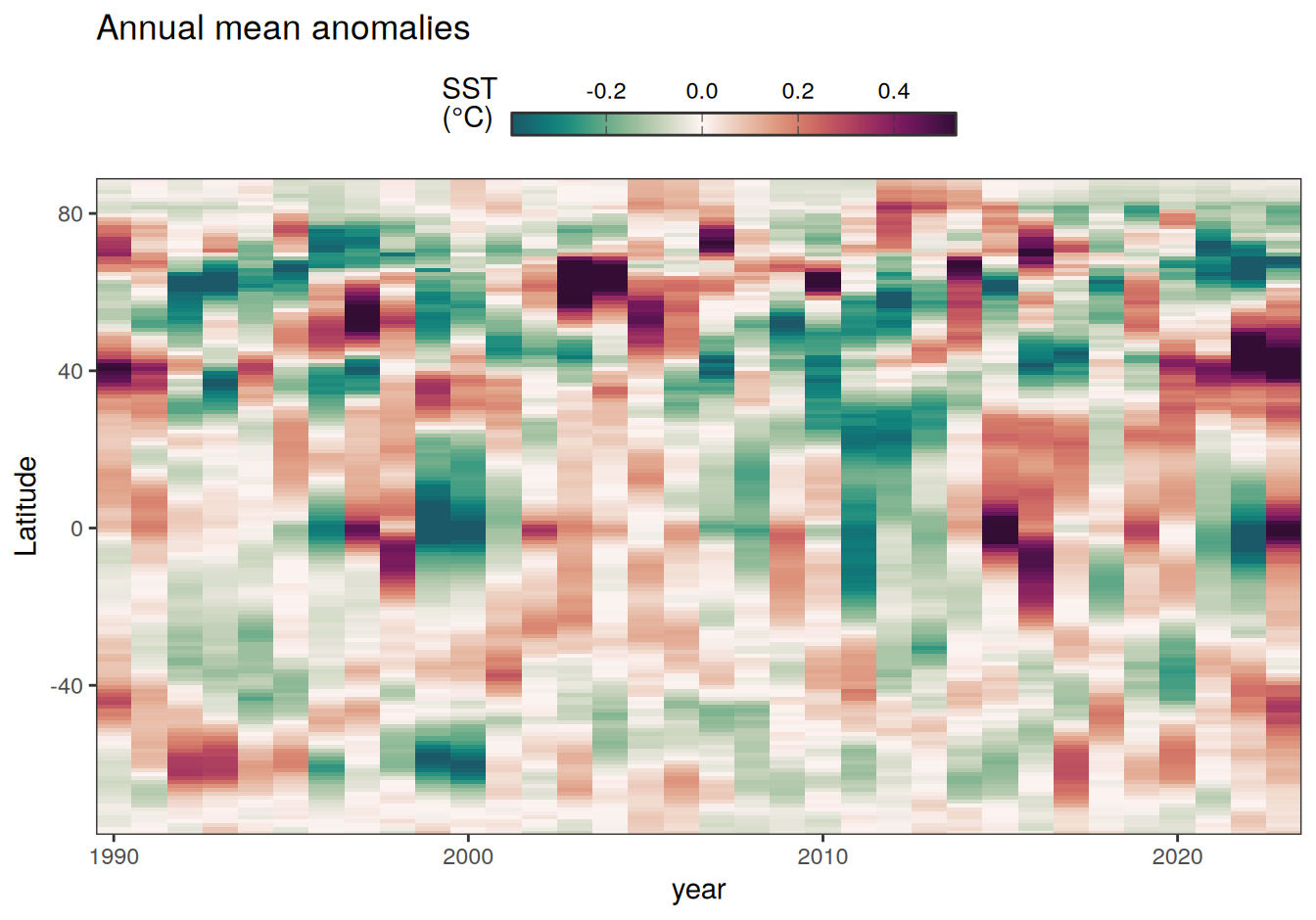
2023 monthly anomalies
pco2_product_hovmoeller_monthly_anomaly <-
pco2_product_hovmoeller_monthly %>%
select(-c(decimal)) %>%
anomaly_determination(lat, month) %>%
filter(!is.na(resid))
pco2_product_hovmoeller_monthly_anomaly <-
pco2_product_hovmoeller_monthly_anomaly %>%
mutate(decimal = year + (month - 1) / 12)
pco2_product_hovmoeller_monthly_anomaly %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(decimal, lat, fill = resid)) +
geom_raster() +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
coord_cartesian(expand = 0) +
labs(title = "Monthly mean anomalies",
y = "Latitude") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")
)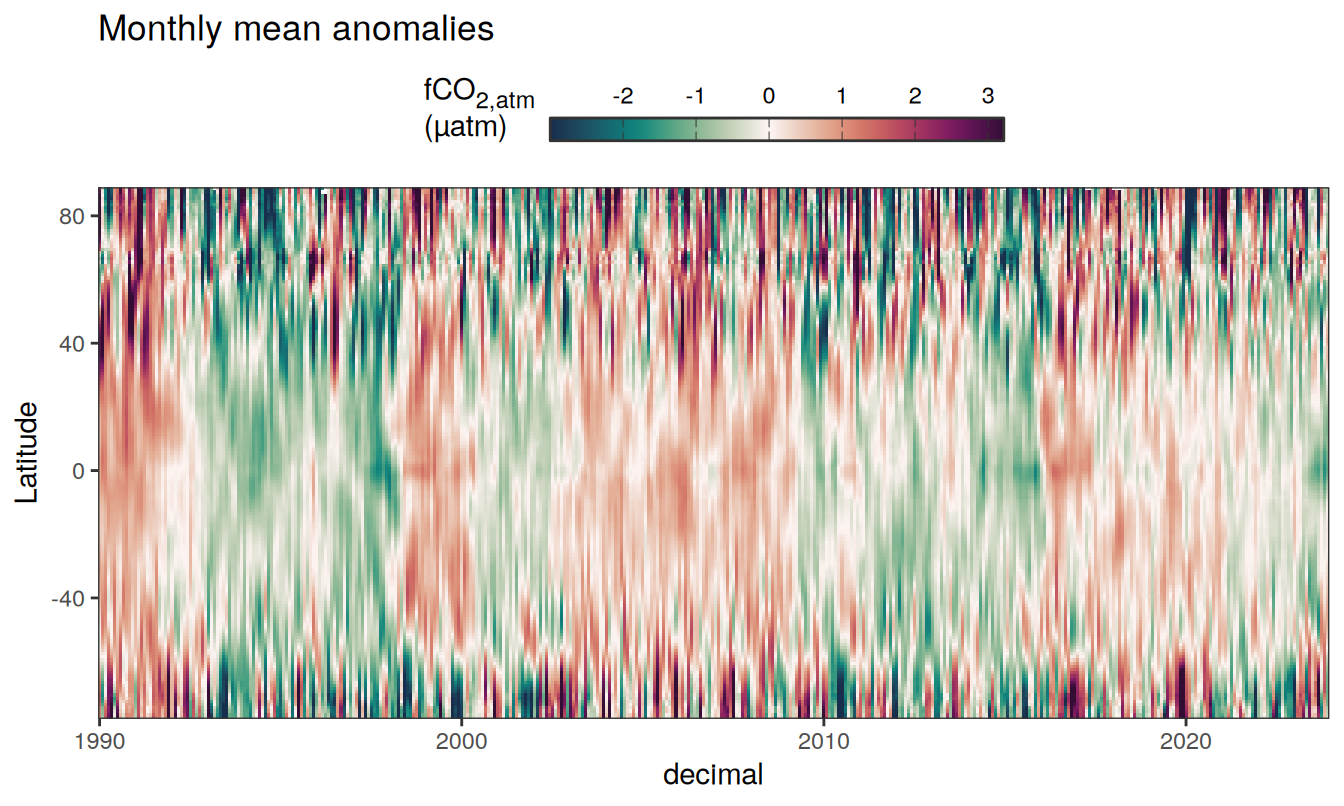
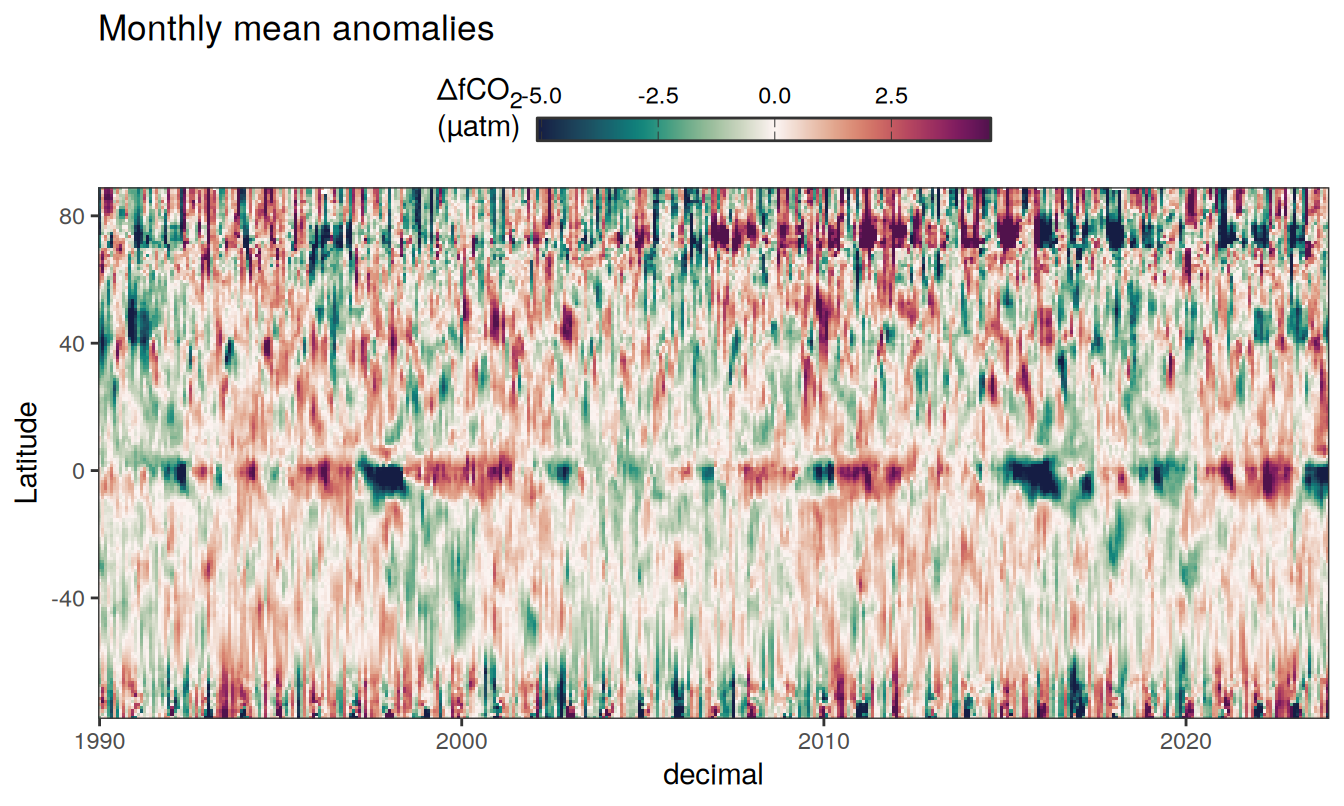
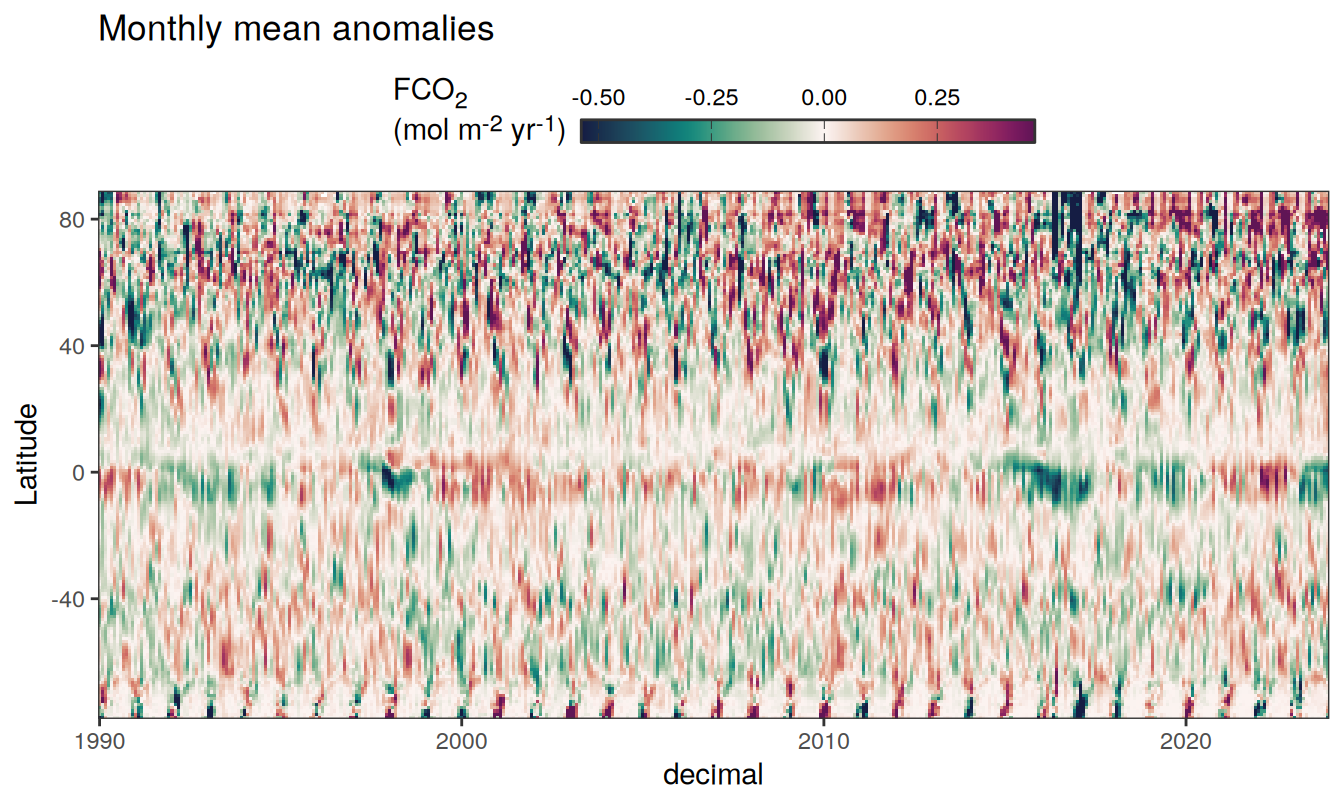
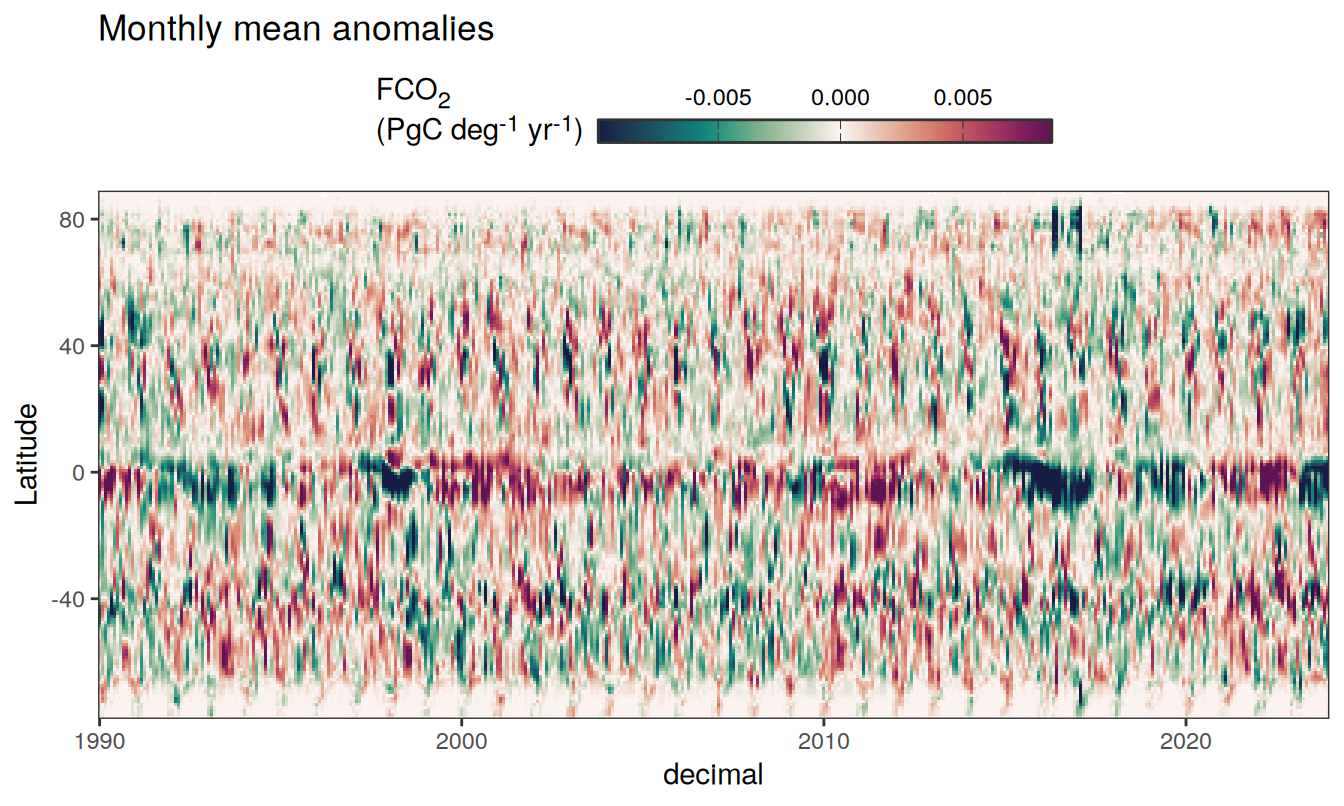
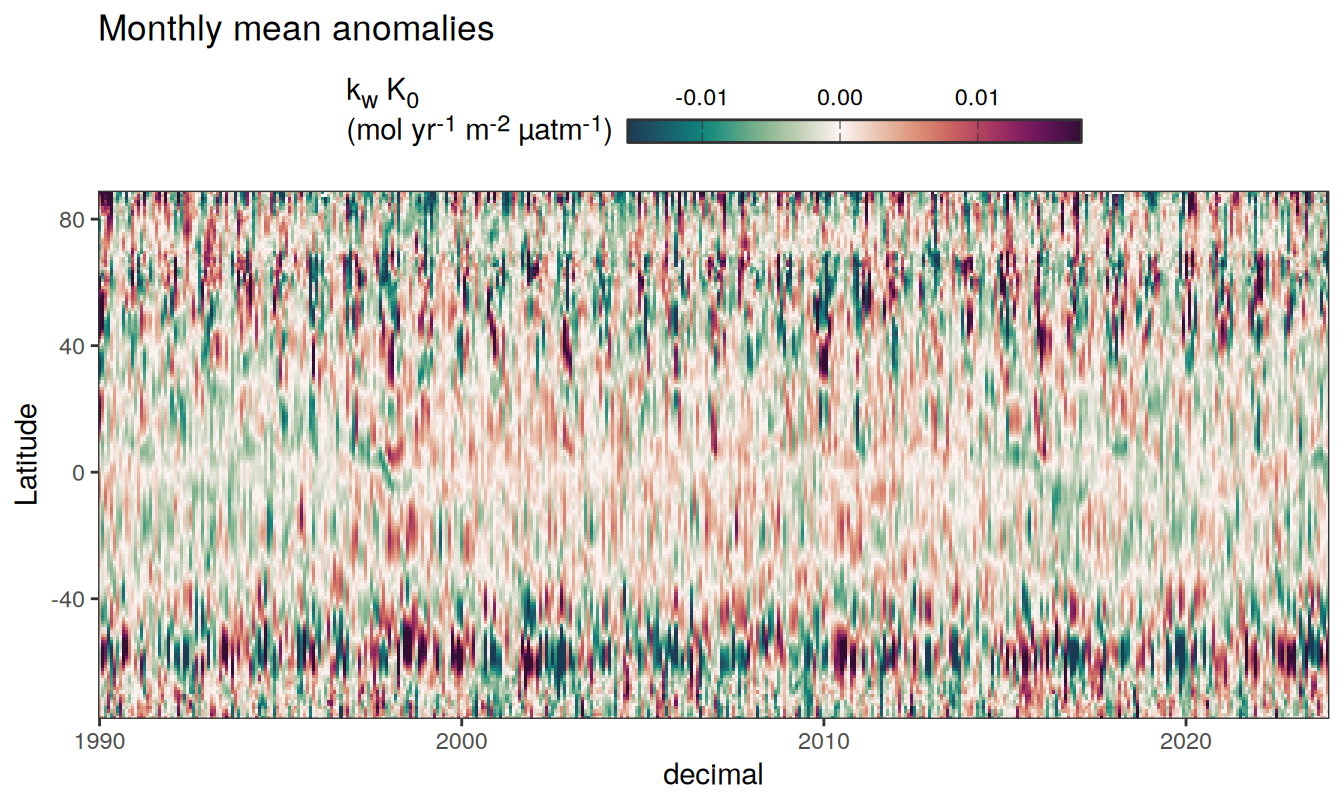
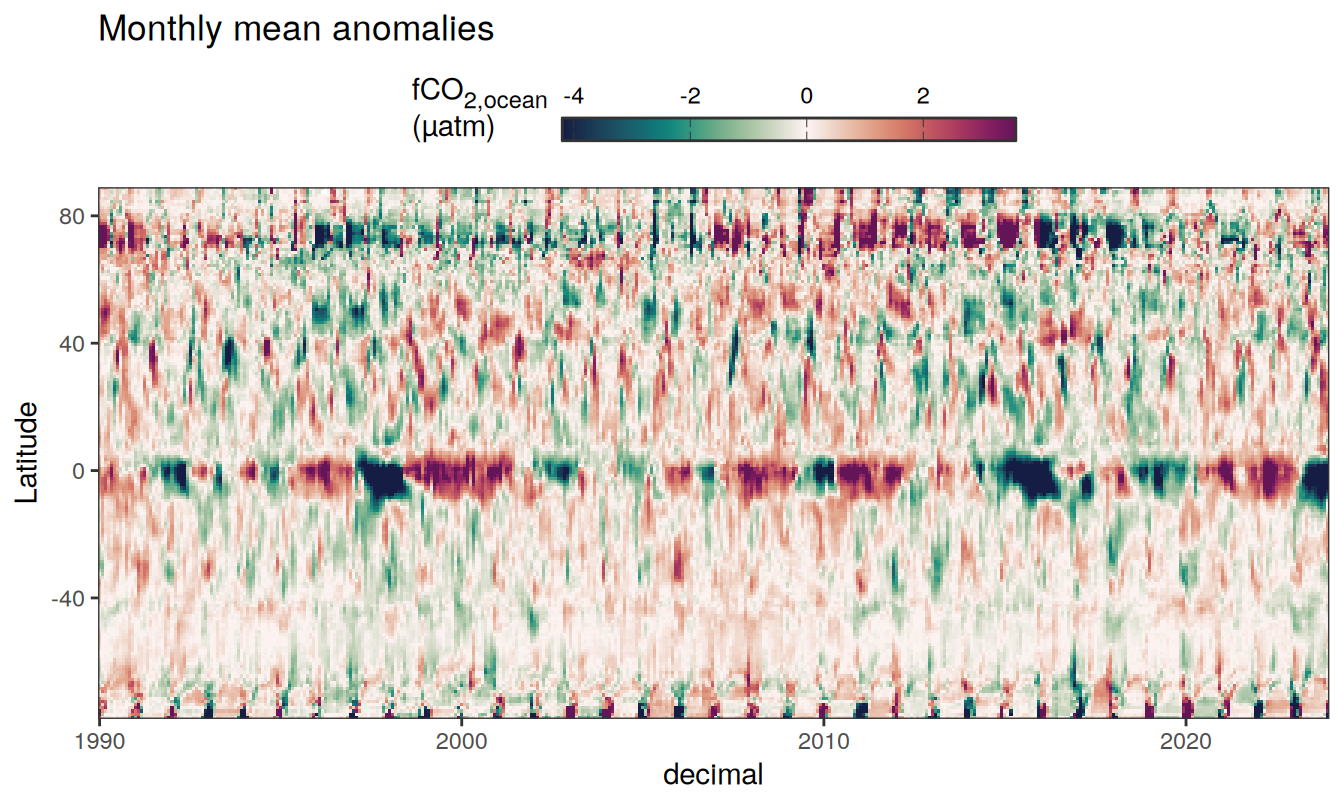
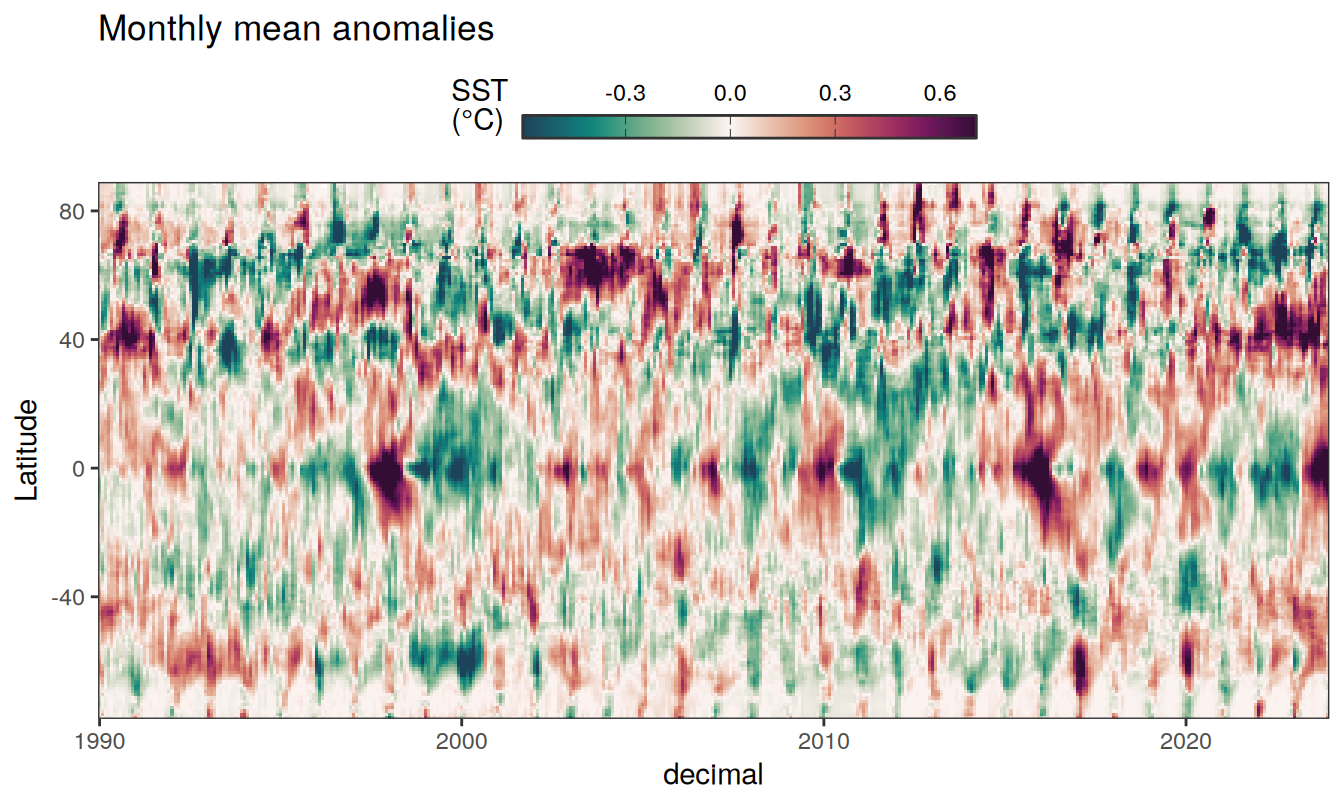
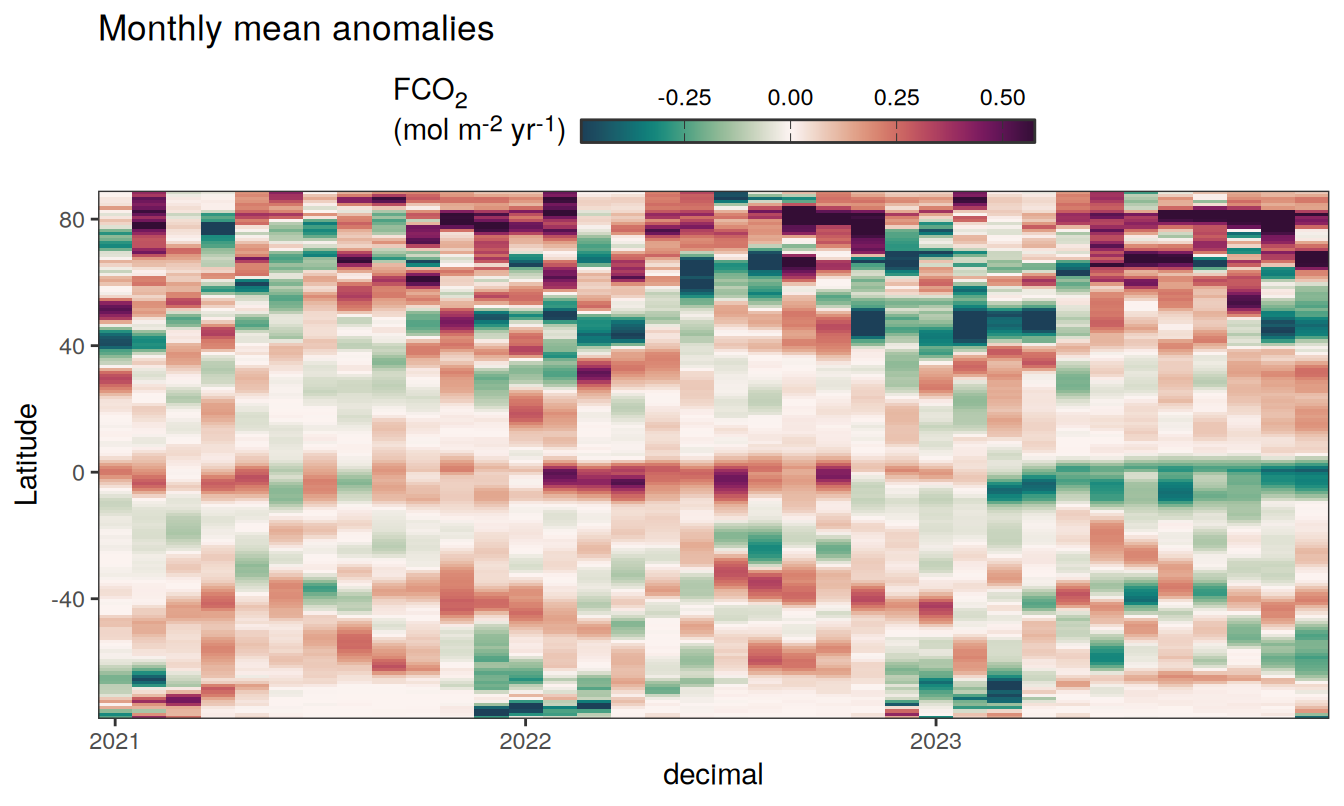
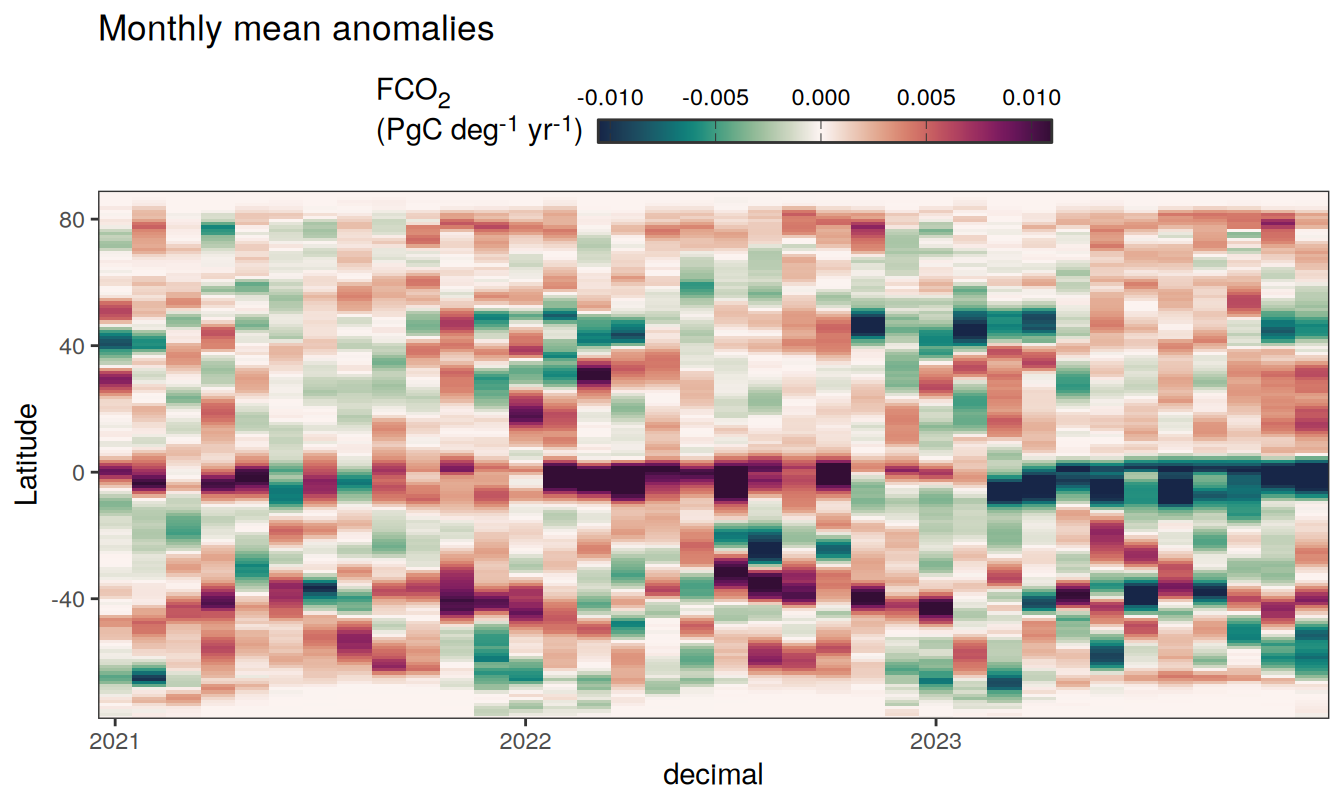
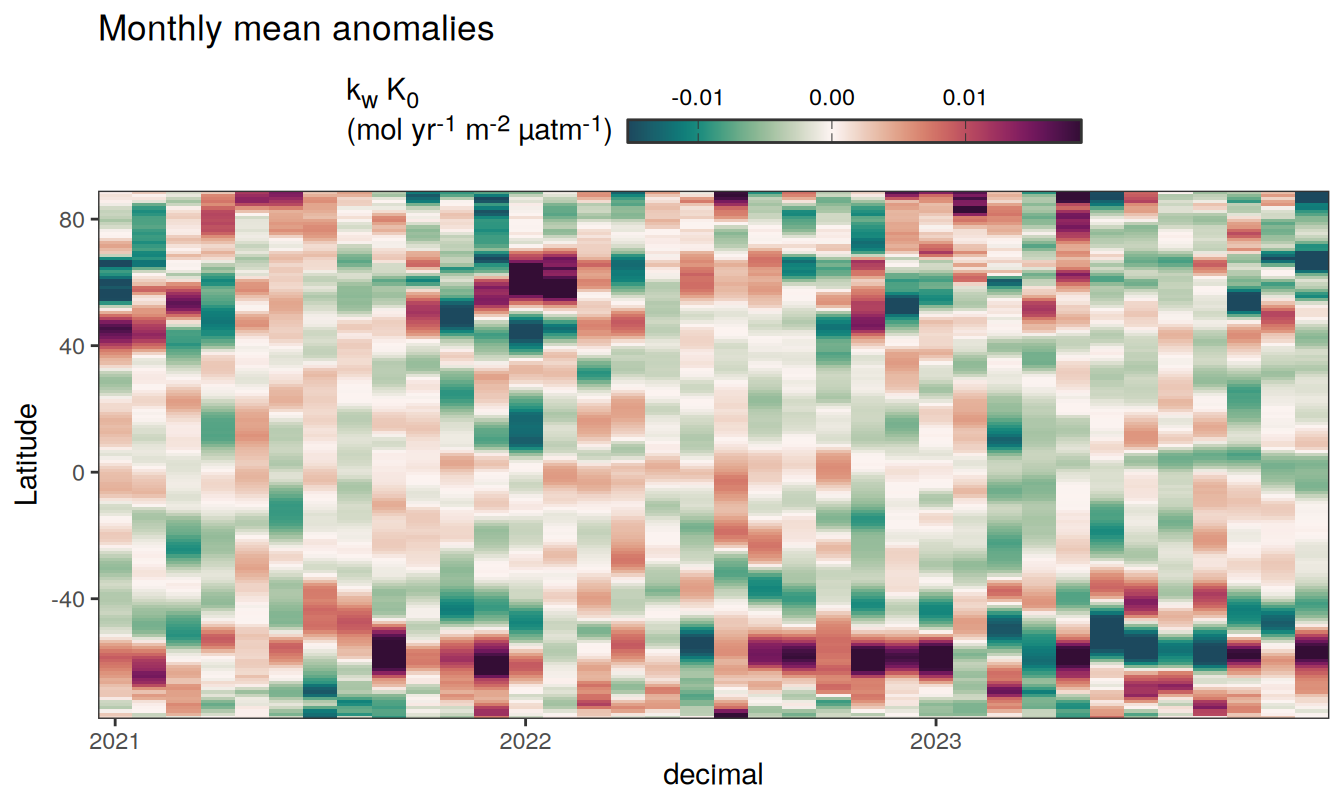
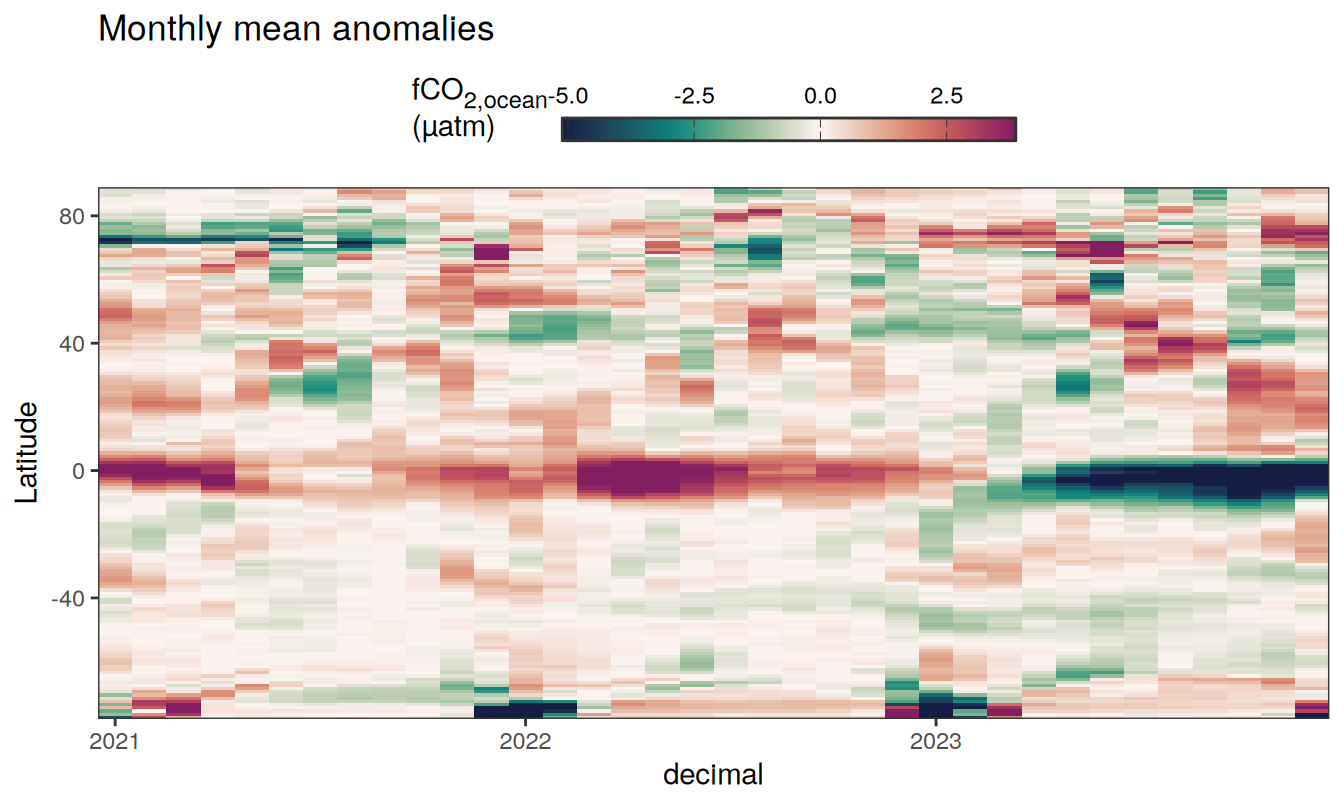
Three years prior 2023
pco2_product_hovmoeller_monthly_anomaly %>%
filter(between(year, 2023-2, 2023)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(decimal, lat, fill = resid)) +
geom_raster() +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
coord_cartesian(expand = 0) +
labs(title = "Monthly mean anomalies",
y = "Latitude") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")
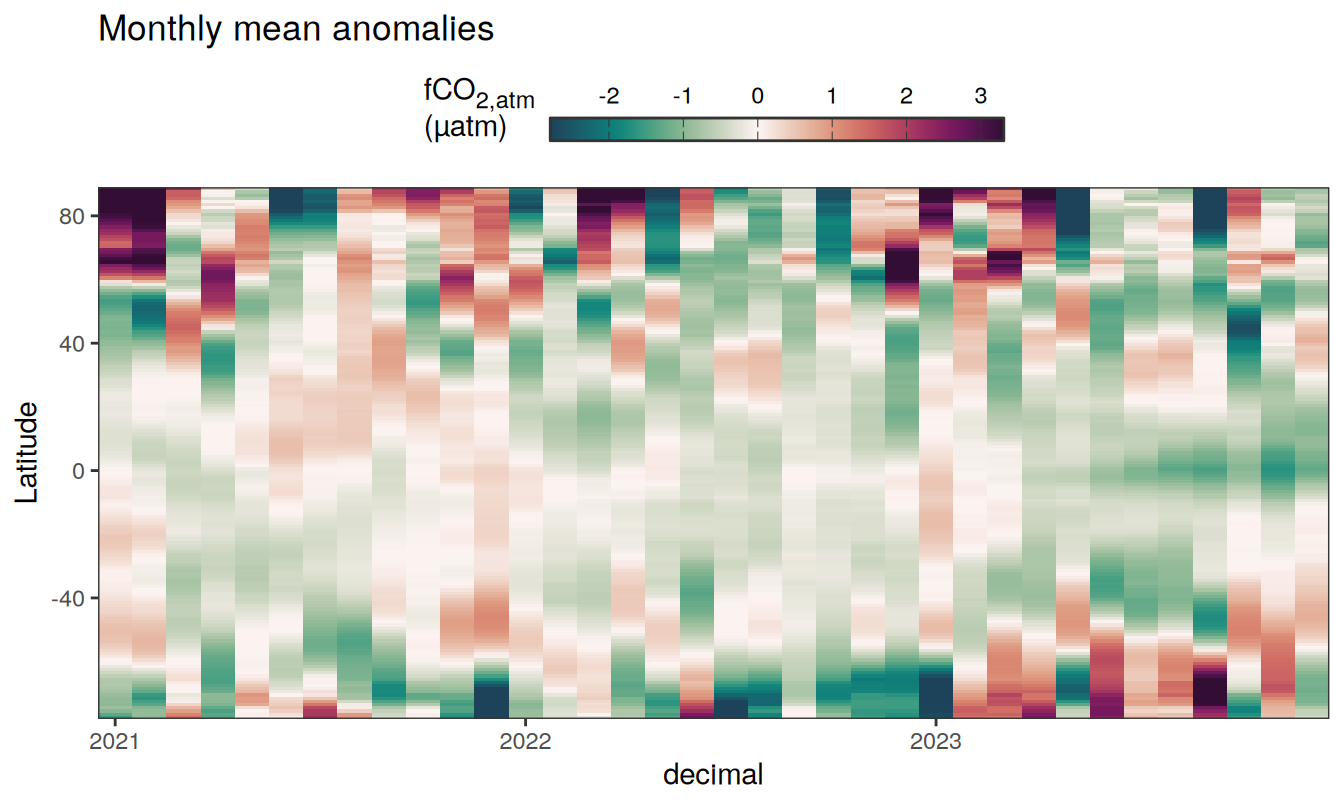
)

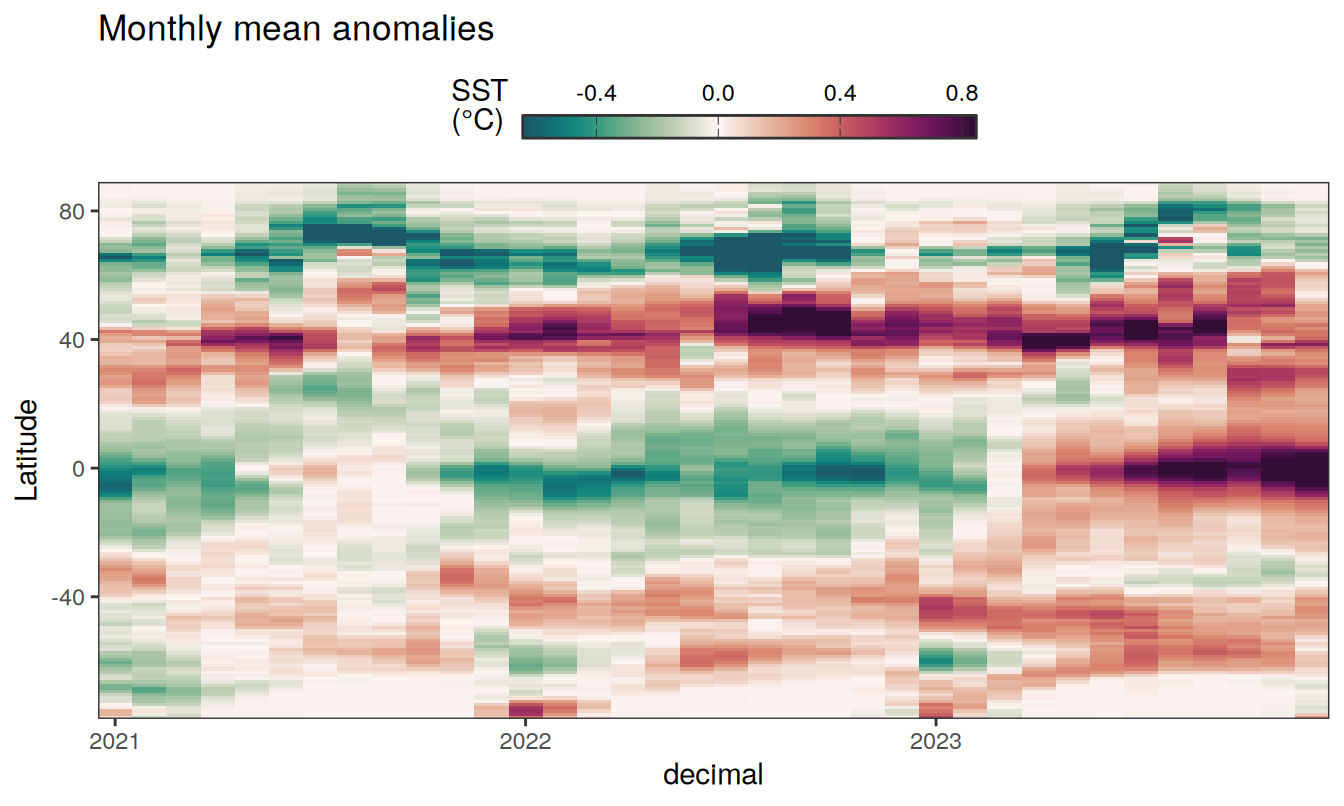
pco2_product_hovmoeller_monthly_anomaly %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_hovmoeller_monthly_anomaly.csv"
)
)
rm(
pco2_product_hovmoeller_annual,
pco2_product_hovmoeller_monthly,
pco2_product_hovmoeller_annual_anomaly,
pco2_product_hovmoeller_monthly_anomaly
)
gc()Regional means and integrals
The following plots show regionally averaged (or integrated) values of each variable as provided through the pCO2 product, as well as the anomalies from the prediction of a linear/quadratic fit.
Anomalies are first presented relative to the predicted annual mean of each year, hence preserving the seasonality. Furthermore, anomalies are presented relative to the predicted monthly mean values, such that the mean seasonality is removed.
2023 absolute values
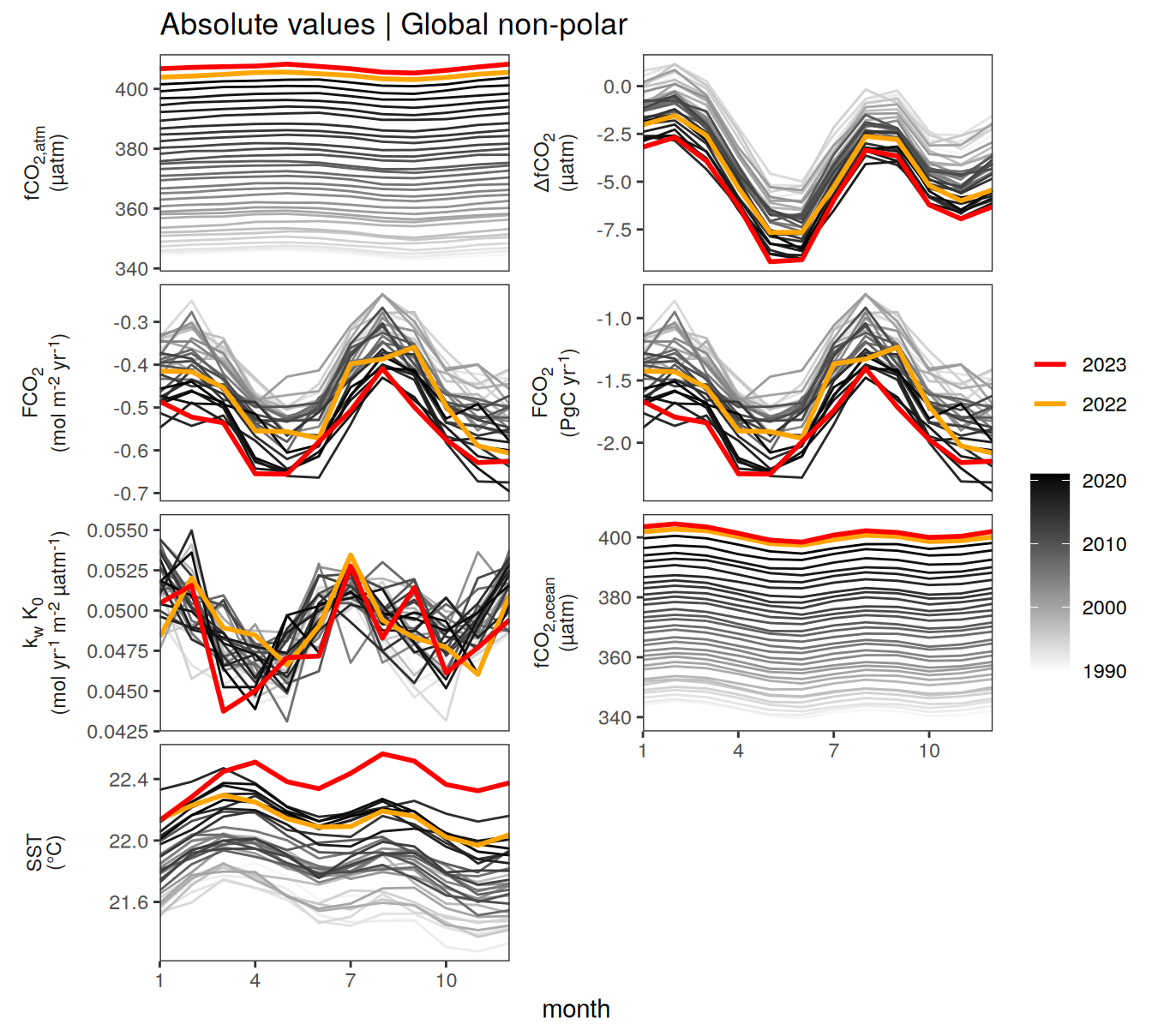
Global non-polar
fig.height <- pco2_product_biome_monthly %>%
distinct(name) %>%
nrow()
fig.height <- (fig.height + 2) * 0.1pco2_product_biome_monthly %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(month, value, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Absolute values | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.title = element_blank(),
axis.title.y = element_blank()
)
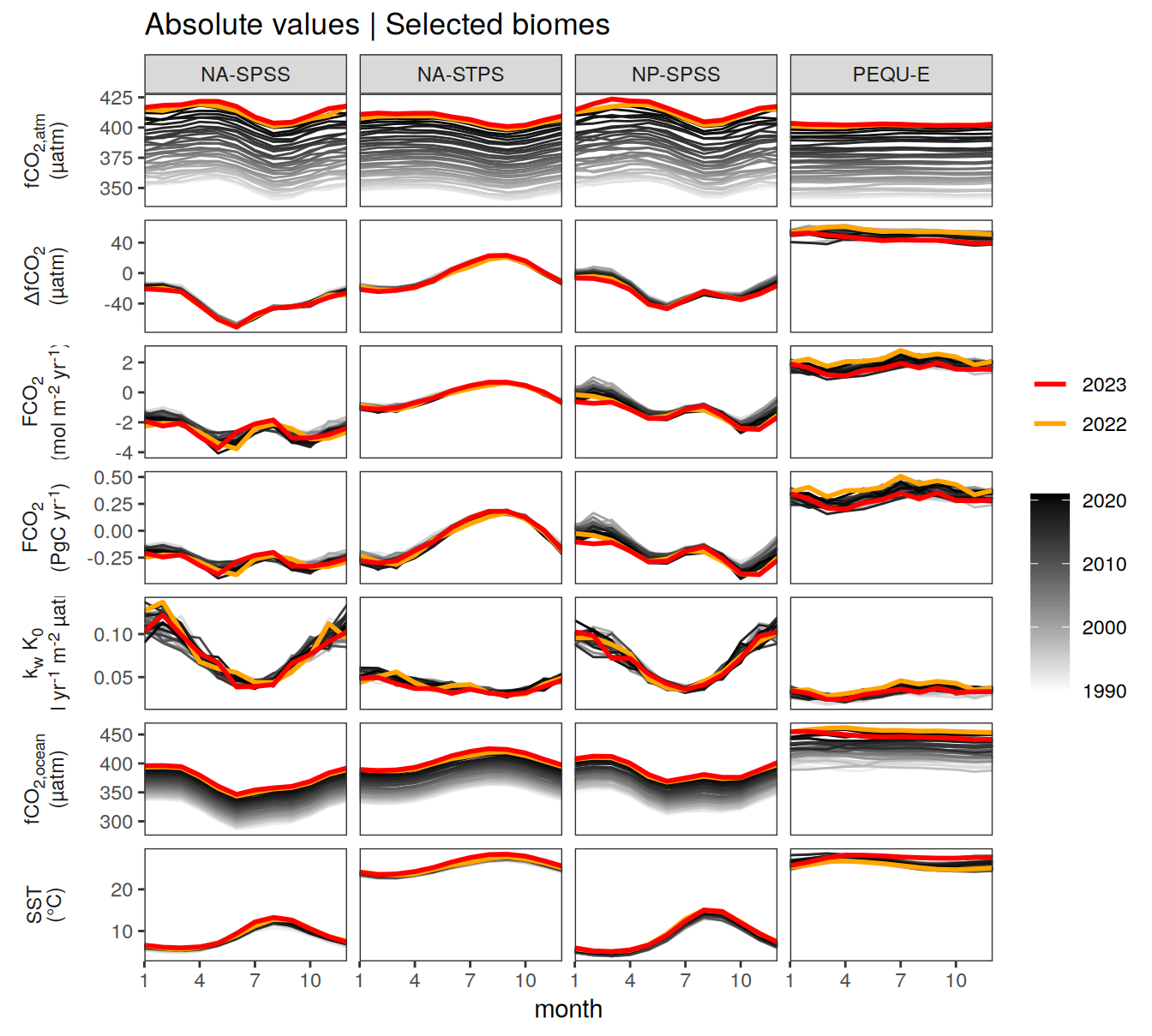
Key biomes
pco2_product_biome_monthly %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, value, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Absolute values | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.title = element_blank(),
axis.title.y = element_blank()
)
pco2_product_biome_monthly %>%
filter(biome %in% key_biomes) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(month, value, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(
data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1
) +
scale_color_manual(
values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = paste("Absolute values |", .x$biome)) +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.title = element_blank(),
axis.title.y = element_blank()
)
)
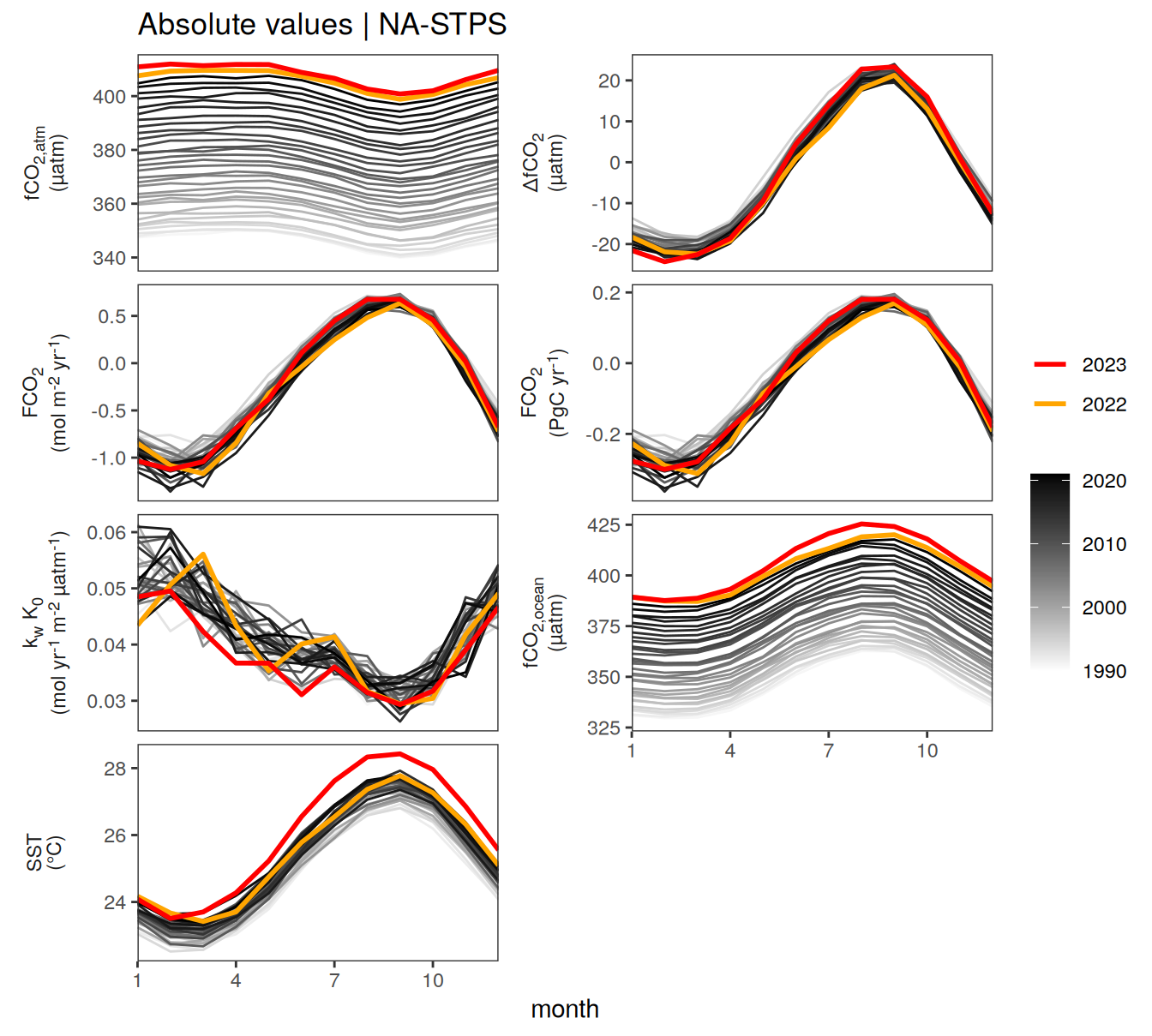
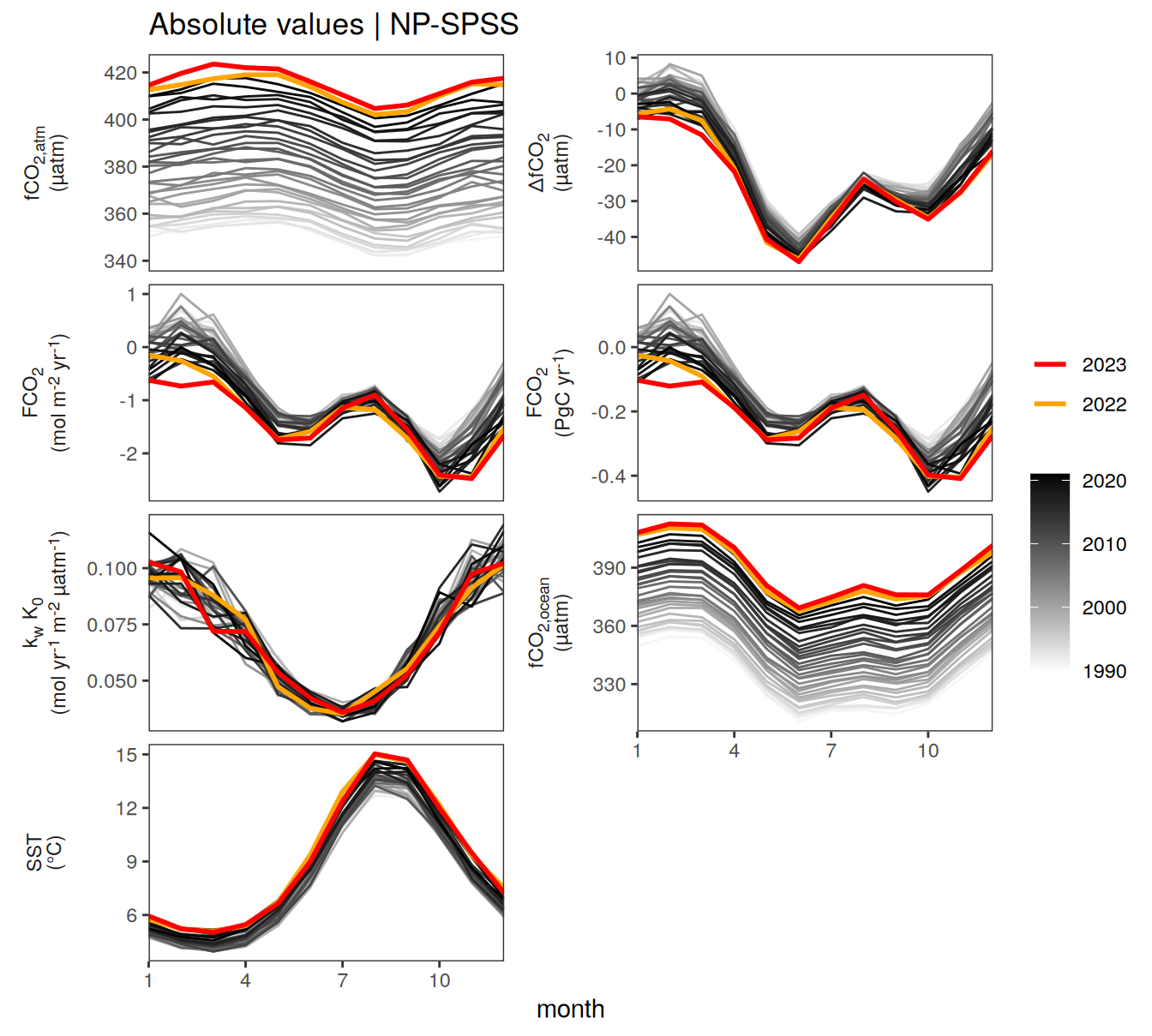
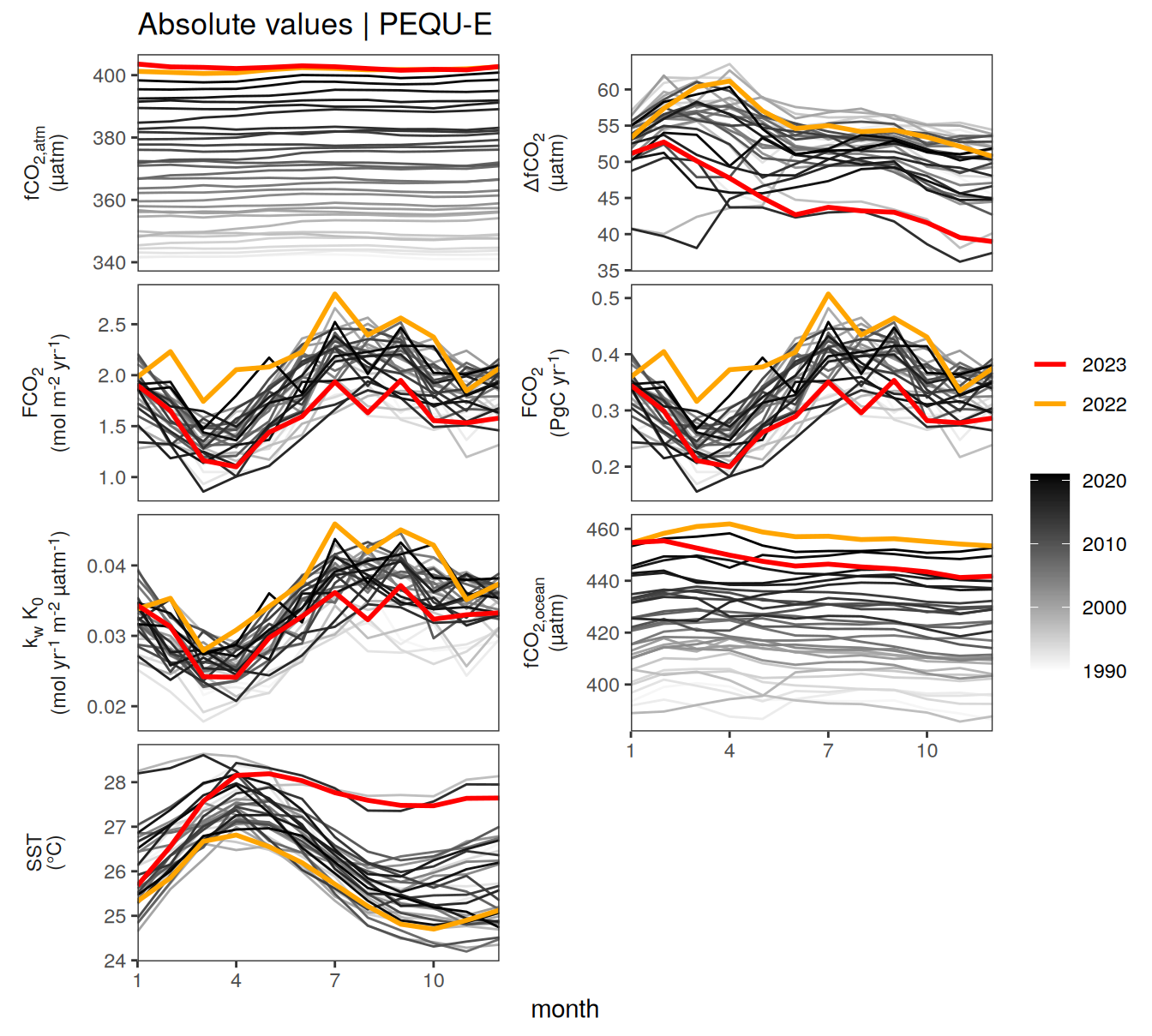
2023 anomalies
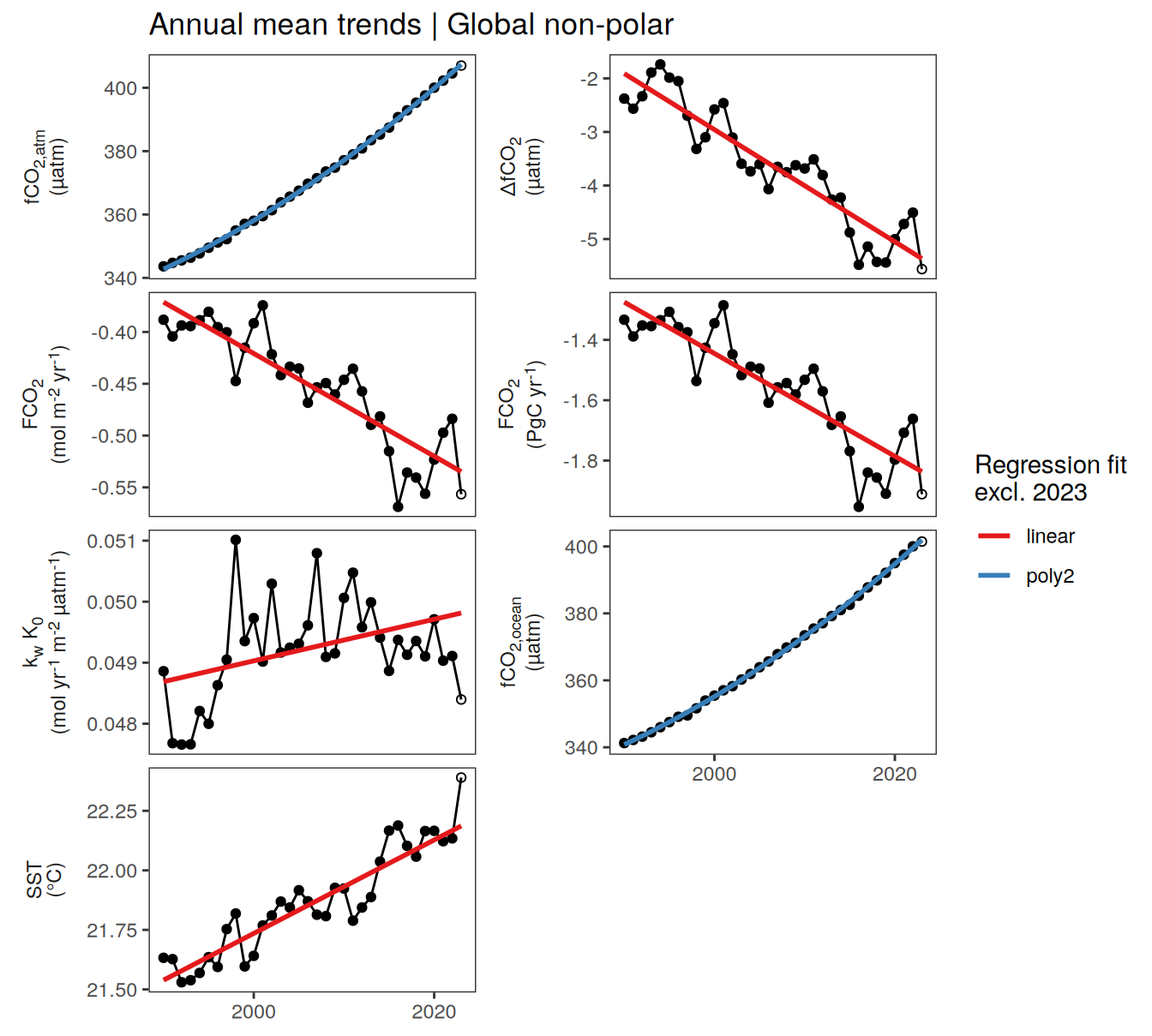
Annual mean trends
pco2_product_biome_annual %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(year, value)) +
geom_path() +
geom_point(data = . %>% filter(year != 2023)) +
geom_point(data = . %>% filter(year == 2023),
shape = 1) +
geom_smooth(data = . %>% filter(year != 2023,
!(name %in% name_quadratic_fit)),
method = "lm",
fullrange = TRUE,
aes(col = "linear"),
se = FALSE) +
geom_smooth(data = . %>% filter(year != 2023,
name %in% name_quadratic_fit),
method = "lm",
fullrange = TRUE,
formula = y ~ poly(x,2),
aes(col = "poly2"),
se = FALSE) +
scale_color_brewer(
palette = "Set1",
name = paste("Regression fit\nexcl.", 2023)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Annual mean trends | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank()
)
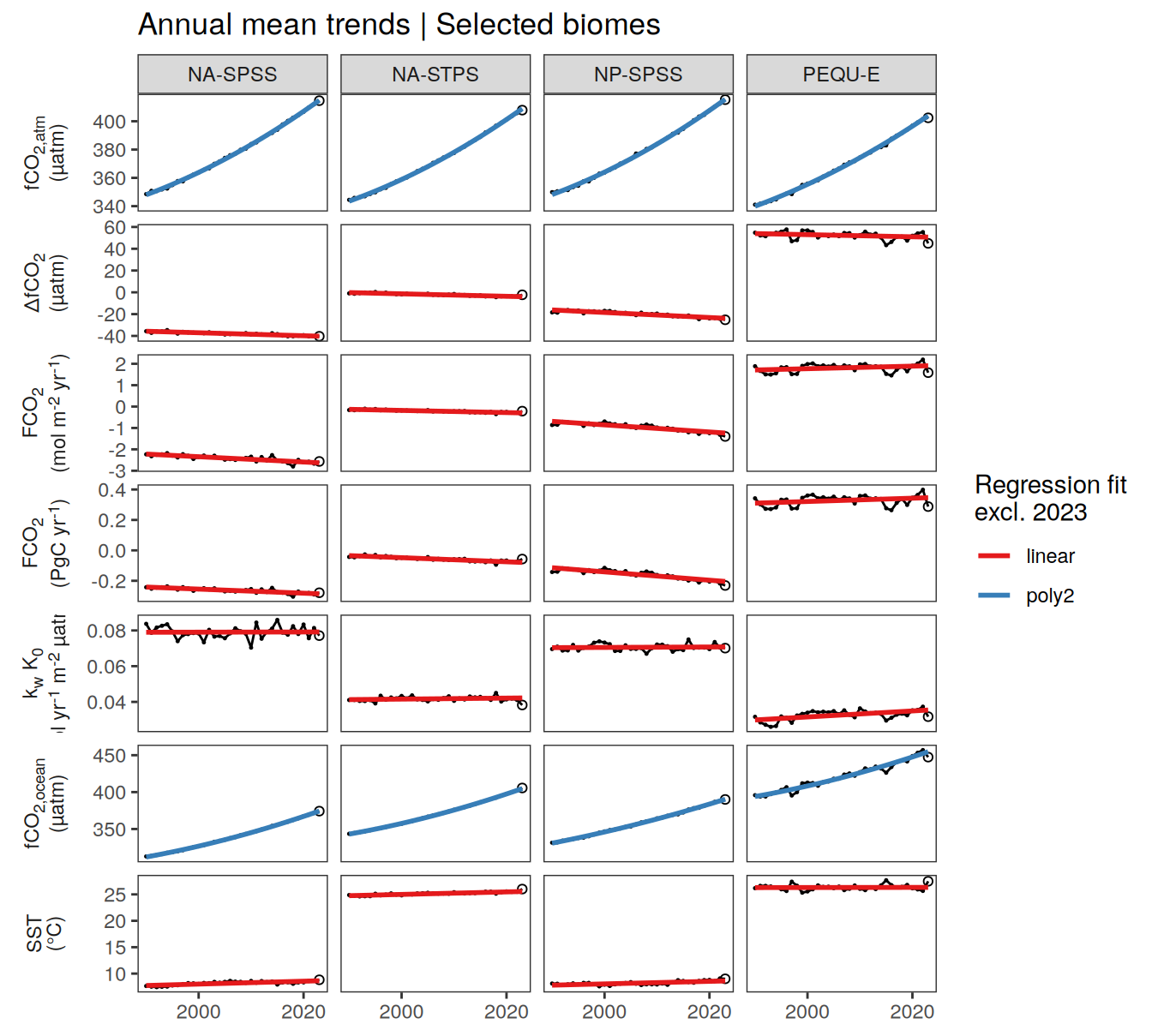
pco2_product_biome_annual %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(year, value)) +
geom_path() +
geom_point(data = . %>% filter(year != 2023),
size = 0.2) +
geom_point(data = . %>% filter(year == 2023),
shape = 1) +
geom_smooth(data = . %>% filter(year != 2023,
!(name %in% name_quadratic_fit)),
method = "lm",
fullrange = TRUE,
aes(col = "linear"),
se = FALSE) +
geom_smooth(data = . %>% filter(year != 2023,
name %in% name_quadratic_fit),
method = "lm",
fullrange = TRUE,
formula = y ~ poly(x,2),
aes(col = "poly2"),
se = FALSE) +
scale_color_brewer(
palette = "Set1",
name = paste("Regression fit\nexcl.", 2023)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Annual mean trends | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank()
)
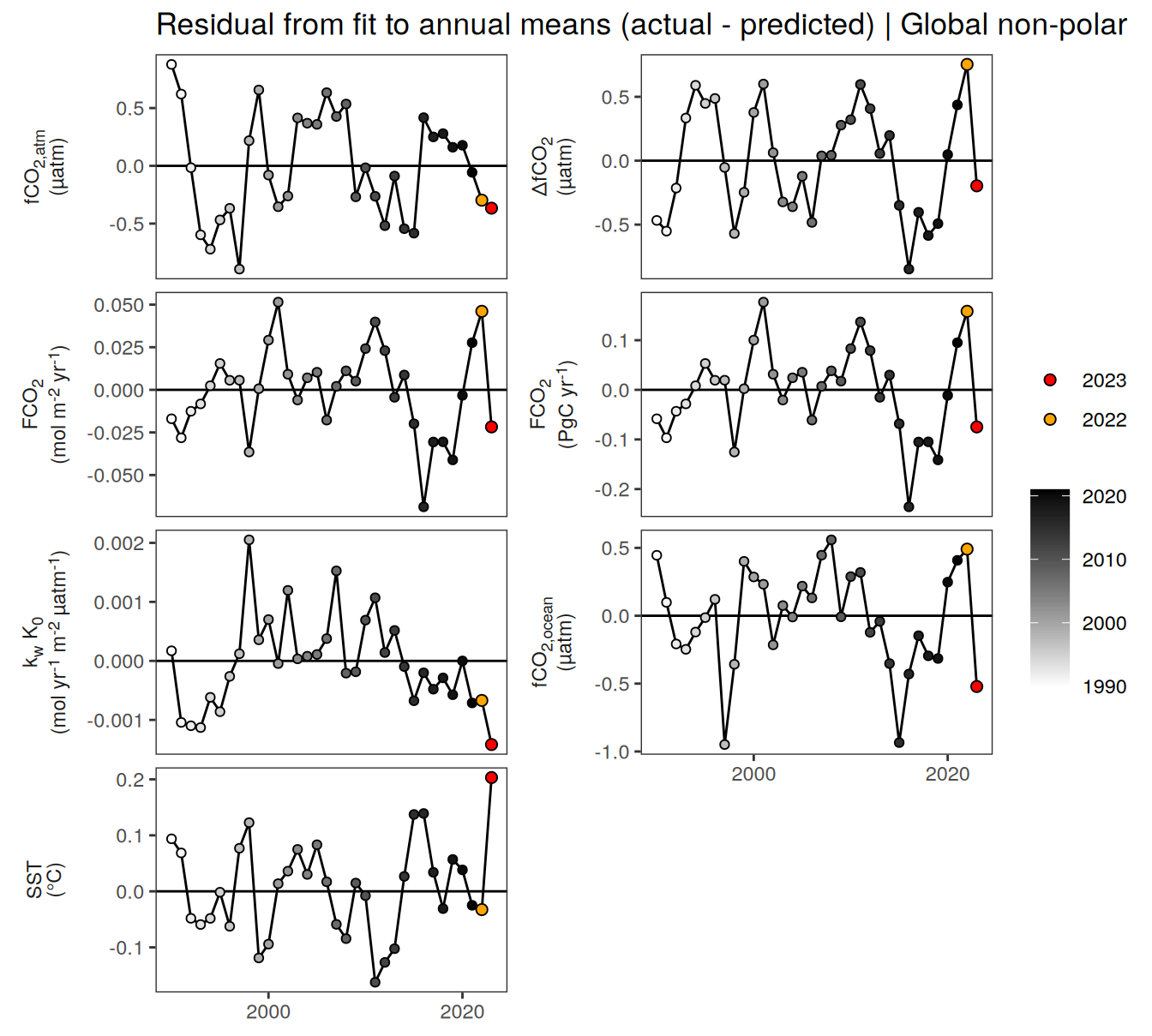
pco2_product_biome_annual_anomaly <-
pco2_product_biome_annual %>%
anomaly_determination(biome)
pco2_product_biome_annual_anomaly %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(year, resid)) +
geom_hline(yintercept = 0) +
geom_path() +
geom_point(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(fill = year),
shape = 21) +
scale_fill_grayC() +
new_scale_fill() +
geom_point(data = . %>% filter(between(year, 2023-1, 2023)),
aes(fill = as.factor(year)),
shape = 21, size = 2) +
scale_fill_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Residual from fit to annual means (actual - predicted) | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank(),
legend.title = element_blank()
)
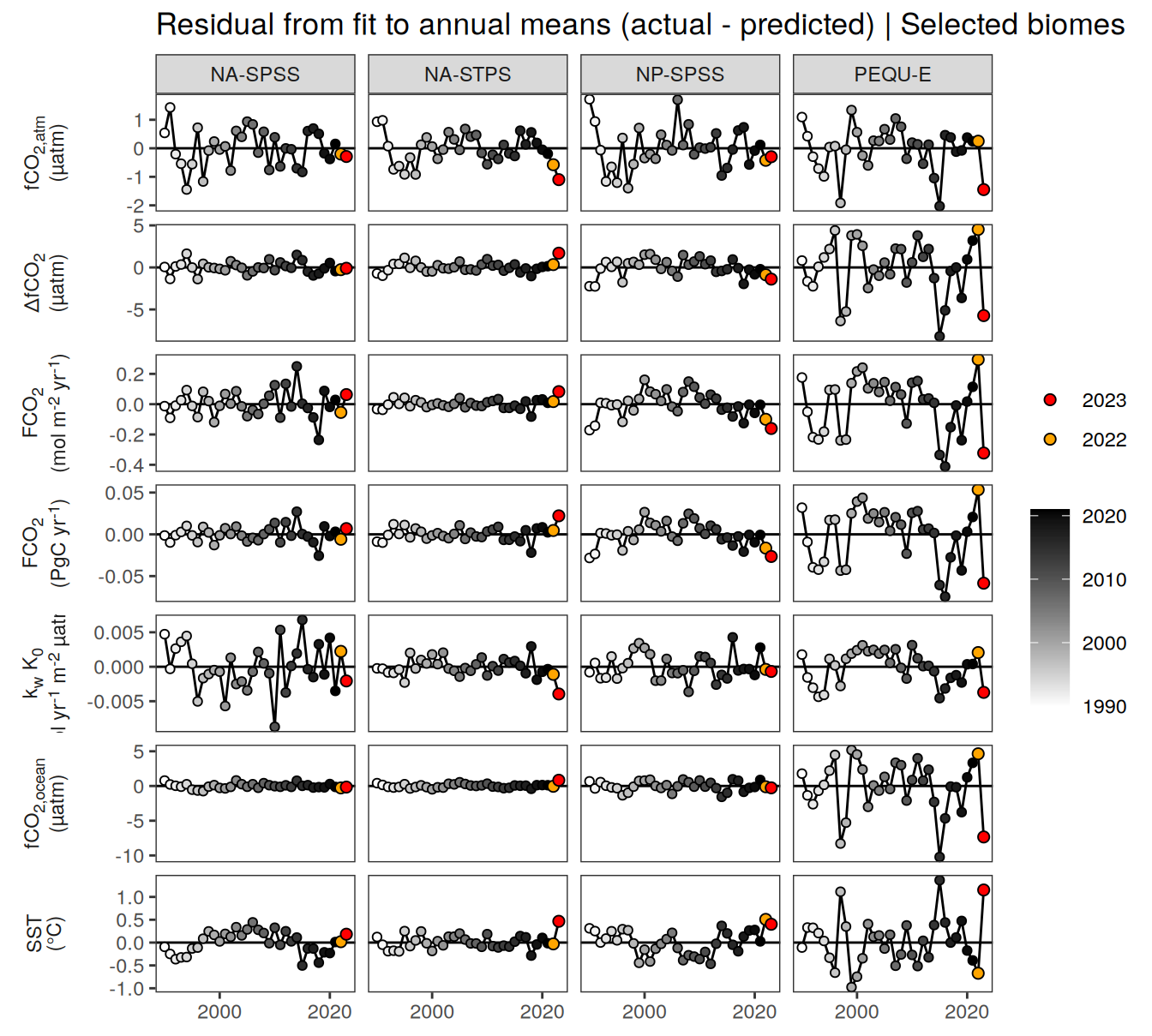
pco2_product_biome_annual_anomaly %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(year, resid)) +
geom_hline(yintercept = 0) +
geom_path() +
geom_point(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(fill = year),
shape = 21) +
scale_fill_grayC() +
new_scale_fill() +
geom_point(data = . %>% filter(between(year, 2023-1, 2023)),
aes(fill = as.factor(year)),
shape = 21, size = 2) +
scale_fill_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Residual from fit to annual means (actual - predicted) | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank(),
legend.title = element_blank()
)
pco2_product_biome_annual_anomaly %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_annual_anomaly.csv"
)
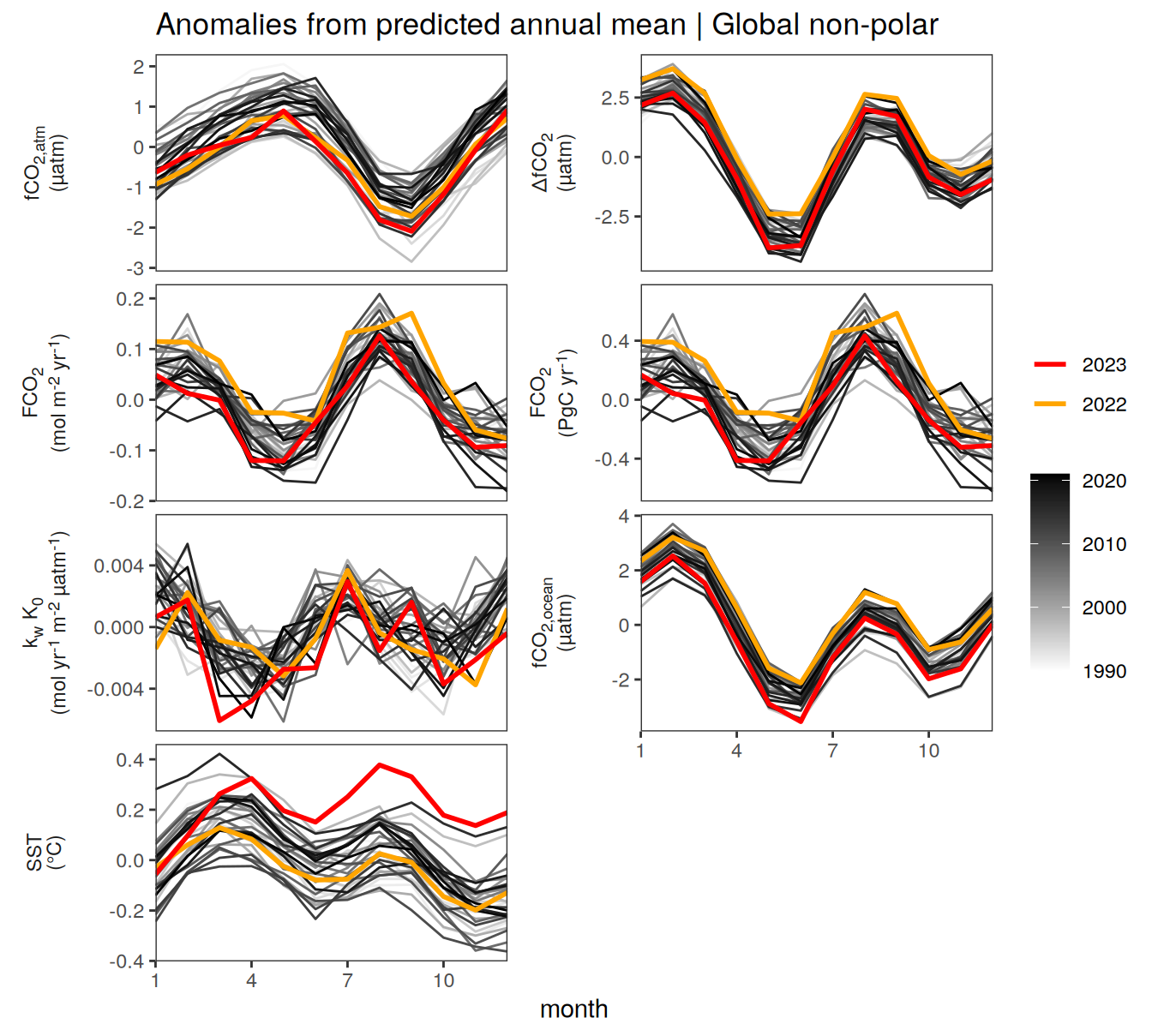
)pco2_product_biome_annual_detrended <-
full_join(pco2_product_biome_monthly,
pco2_product_biome_annual_anomaly %>% select(-c(value, resid))) %>%
mutate(resid = value - fit)
pco2_product_biome_annual_detrended %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted annual mean | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)
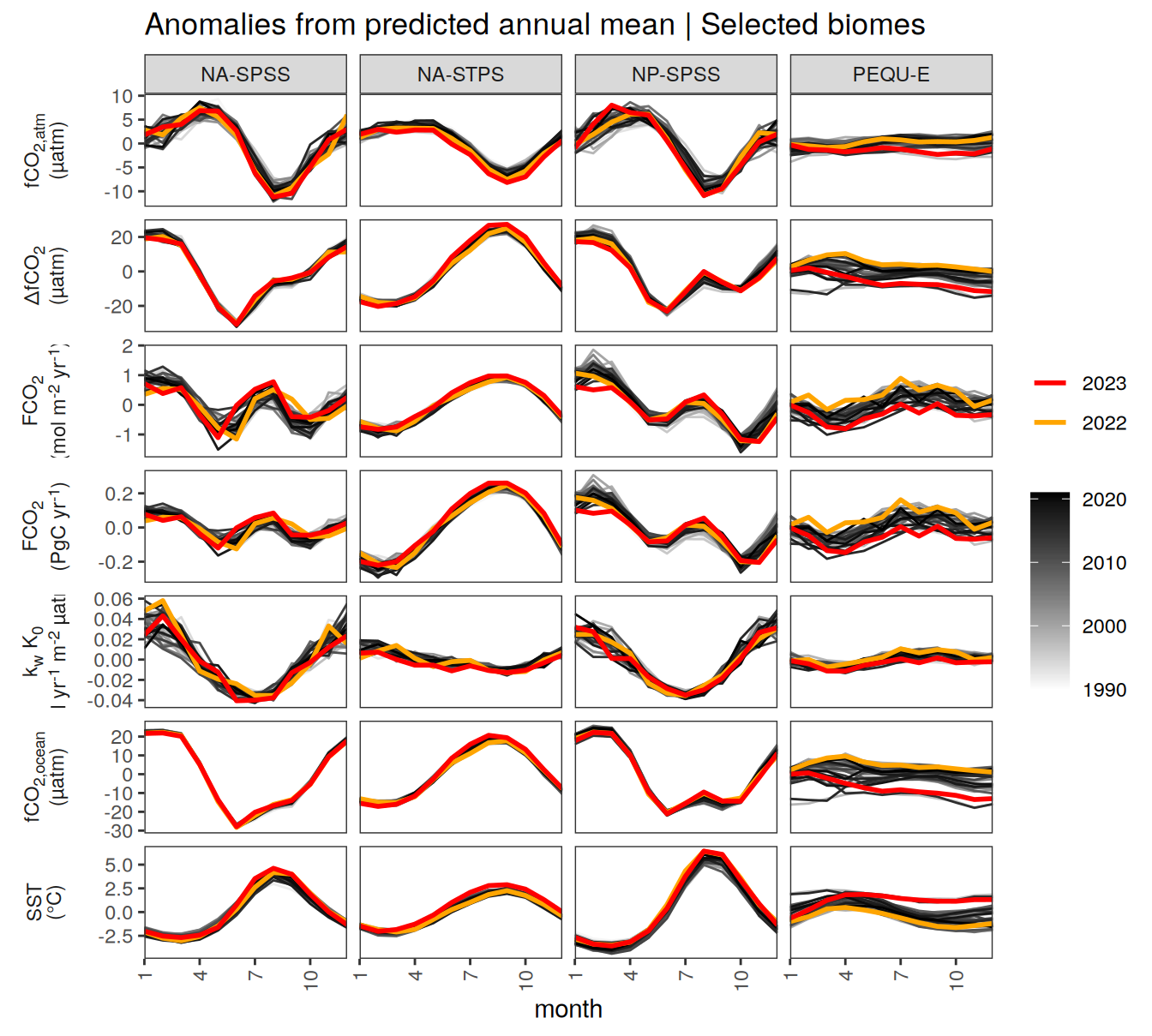
pco2_product_biome_annual_detrended %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted annual mean | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank(),
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)
)
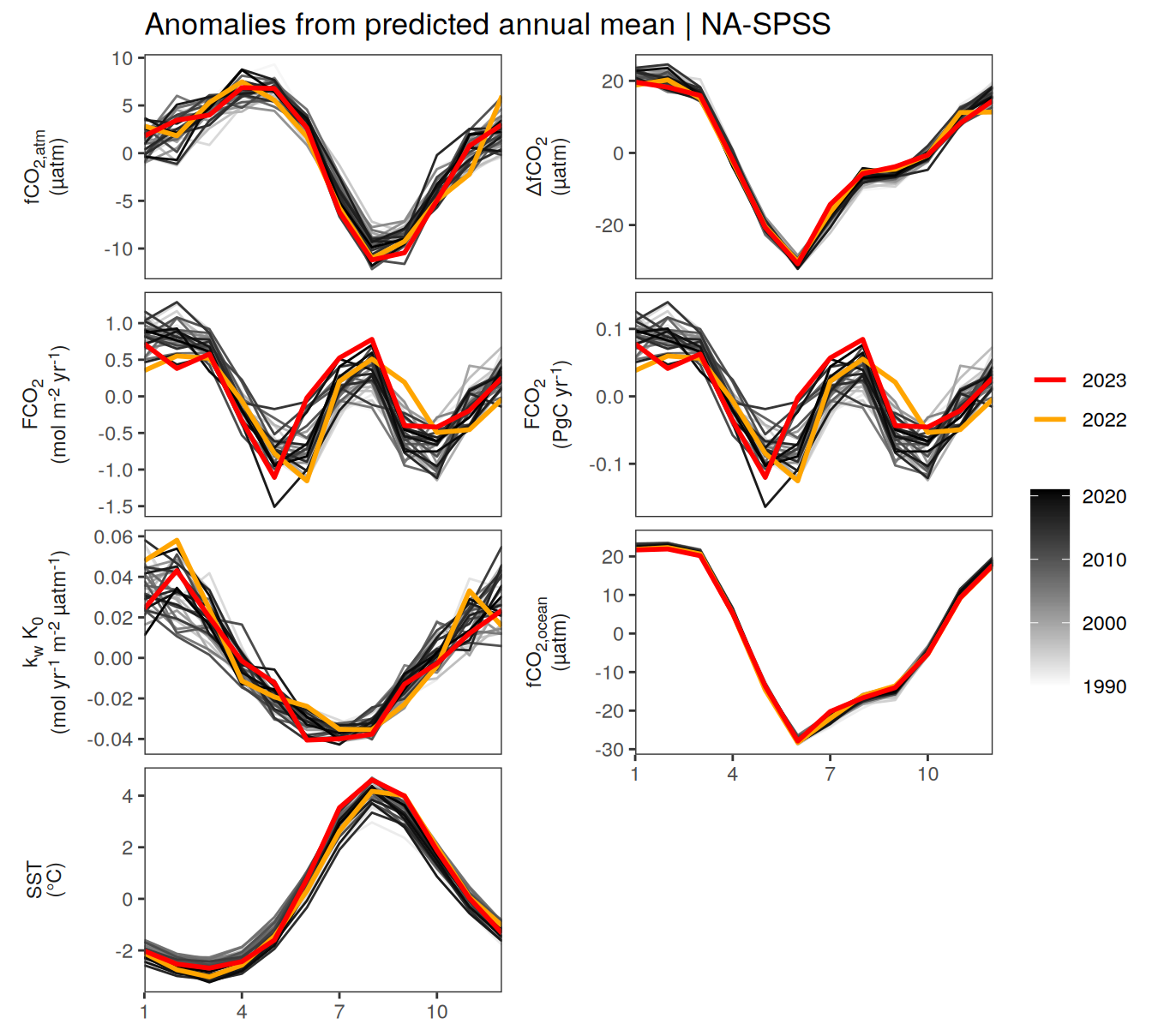
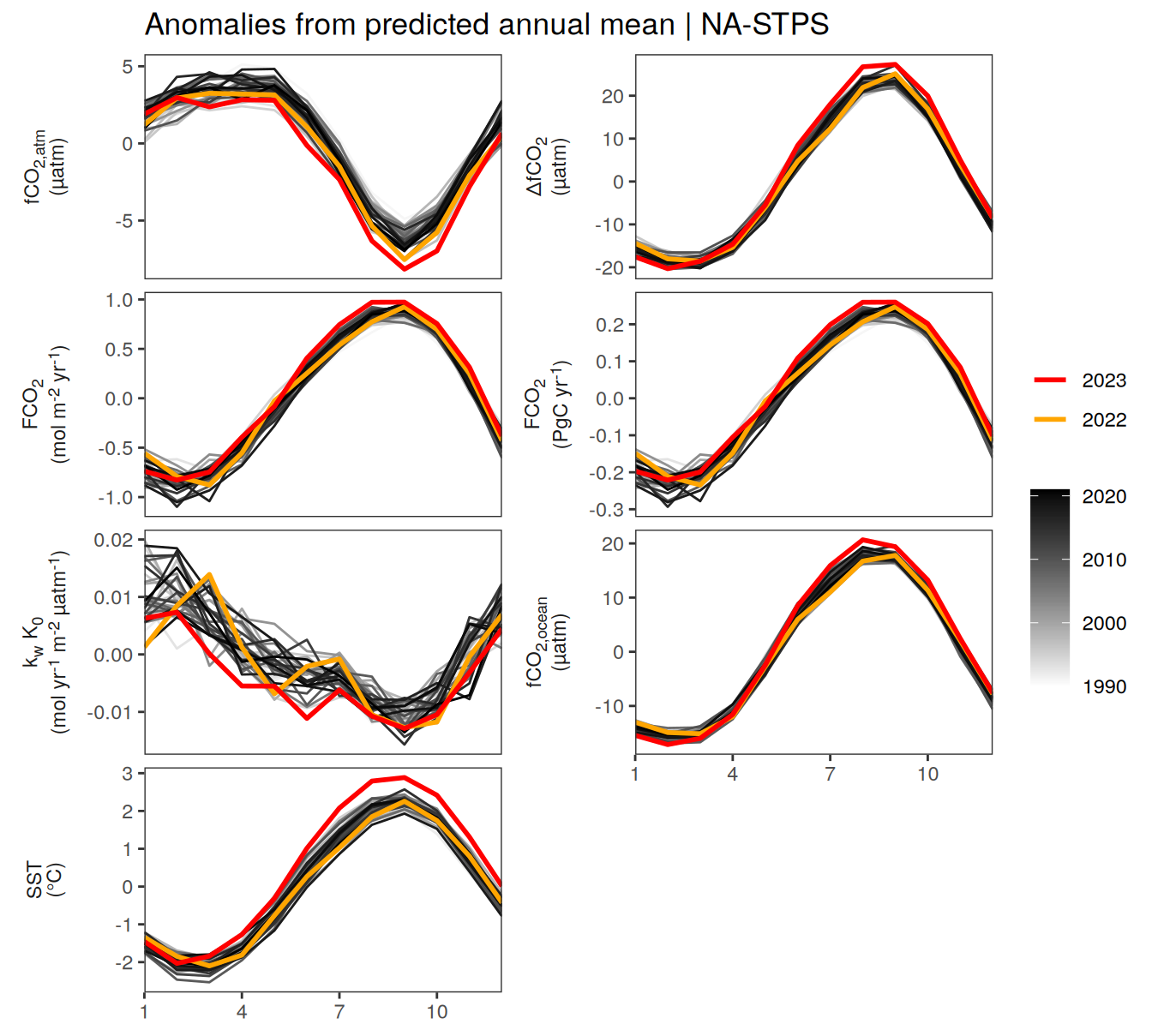
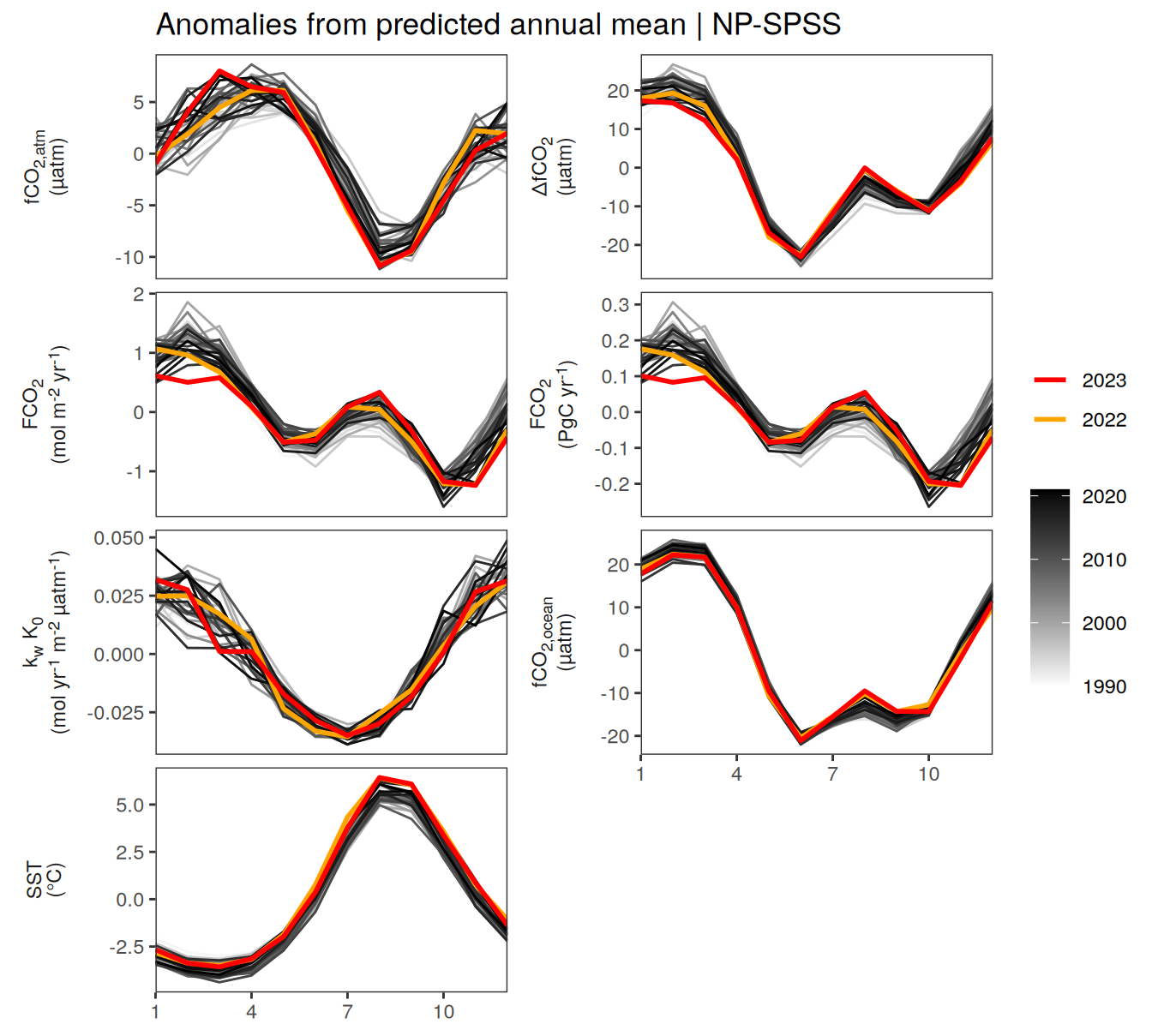
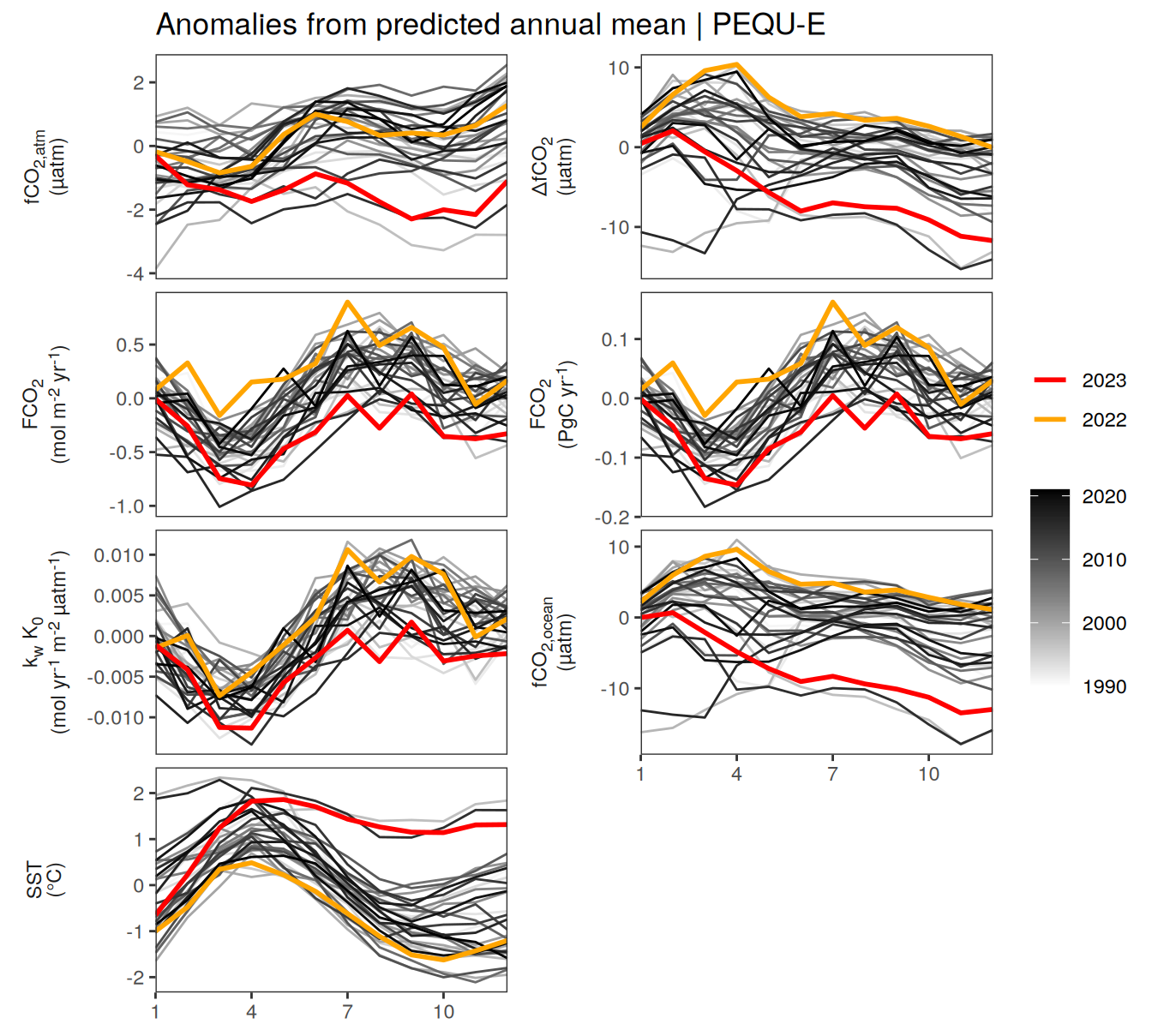
pco2_product_biome_annual_detrended %>%
filter(biome %in% key_biomes) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(
data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1
) +
scale_color_manual(
values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = paste("Anomalies from predicted annual mean |", .x$biome)) +
facet_wrap(
name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank(),
legend.title = element_blank()
)
)
pco2_product_biome_annual_detrended %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_annual_detrended.csv"
)
)Monthly mean trends
# pco2_product_biome_monthly %>%
# filter(biome %in% "Global non-polar") %>%
# mutate(month = as.factor(month)) %>%
# ggplot(aes(year, value, col = month)) +
# # geom_point() +
# geom_smooth(data = . %>% filter(year != 2023,
# !(name %in% name_quadratic_fit)),
# method = "lm",
# se = FALSE,
# fullrange = TRUE) +
# geom_smooth(
# data = . %>% filter(year != 2023,
# name %in% name_quadratic_fit),
# method = "lm",
# fullrange = TRUE,
# formula = y ~ x + I(x ^ 2),
# se = FALSE
# ) +
# scale_color_scico_d(palette = "romaO",
# name = paste("Regression fit\nexcl.", 2023)) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Monthly mean trends | Global non-polar") +
# facet_wrap(name ~ .,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# strip.position = "left",
# ncol = 2) +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank()
# )
#
# pco2_product_biome_monthly %>%
# filter(biome %in% key_biomes) %>%
# mutate(month = as.factor(month)) %>%
# ggplot(aes(year, value, col = month)) +
# # geom_point() +
# geom_smooth(data = . %>% filter(year != 2023,
# !(name %in% name_quadratic_fit)),
# method = "lm",
# fullrange = TRUE,
# se = FALSE) +
# geom_smooth(
# data = . %>% filter(year != 2023,
# name %in% name_quadratic_fit),
# method = "lm",
# fullrange = TRUE,
# formula = y ~ x + I(x ^ 2),
# se = FALSE
# ) +
# scale_color_scico_d(palette = "romaO",
# name = paste("Regression fit\nexcl.", 2023)) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Monthly mean trends | Selected biomes") +
# facet_grid(name ~ biome,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# switch = "y") +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank()
# )
pco2_product_biome_monthly_anomaly <-
pco2_product_biome_monthly %>%
anomaly_determination(biome, month)
# pco2_product_biome_monthly_anomaly %>%
# filter(biome %in% "Global non-polar") %>%
# group_split(month) %>%
# head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, resid)) +
# geom_hline(yintercept = 0) +
# geom_path() +
# geom_point(
# data = . %>% filter(!between(year, 2023-1, 2023)),
# aes(fill = year),
# shape = 21
# ) +
# scale_fill_grayC() +
# new_scale_fill() +
# geom_point(
# data = . %>% filter(between(year, 2023-1, 2023)),
# aes(fill = as.factor(year)),
# shape = 21,
# size = 2
# ) +
# scale_fill_manual(
# values = c("orange", "red"),
# guide = guide_legend(reverse = TRUE,
# order = 1)
# ) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Residual from fit to monthly means (actual - predicted) | Global non-polar",
# subtitle = paste("Month:", .x$month)) +
# facet_wrap(
# name ~ .,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# strip.position = "left",
# ncol = 2
# ) +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank(),
# legend.title = element_blank()
# )
# )
#
# pco2_product_biome_monthly_anomaly %>%
# filter(biome %in% key_biomes) %>%
# group_split(month) %>%
# head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, resid)) +
# geom_hline(yintercept = 0) +
# geom_path() +
# geom_point(
# data = . %>% filter(!between(year, 2023-1, 2023)),
# aes(fill = year),
# shape = 21
# ) +
# scale_fill_grayC() +
# new_scale_fill() +
# geom_point(
# data = . %>% filter(between(year, 2023-1, 2023)),
# aes(fill = as.factor(year)),
# shape = 21,
# size = 2
# ) +
# scale_fill_manual(
# values = c("orange", "red"),
# guide = guide_legend(reverse = TRUE,
# order = 1)
# ) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Residual from fit to monthly means (actual - predicted) | Selected biomes",
# subtitle = paste("Month:", .x$month)) +
# facet_grid(
# name ~ biome,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# switch = "y"
# ) +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank(),
# legend.title = element_blank()
# )
# )pco2_product_biome_monthly_anomaly %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_monthly_anomaly.csv"
)
)Global non-polar
pco2_product_biome_monthly_detrended <-
full_join(pco2_product_biome_monthly,
pco2_product_biome_monthly_anomaly %>% select(-c(value, resid))) %>%
mutate(resid = value - fit)
pco2_product_biome_monthly_detrended %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted monthly mean | Global non-polar") +
facet_wrap(
name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)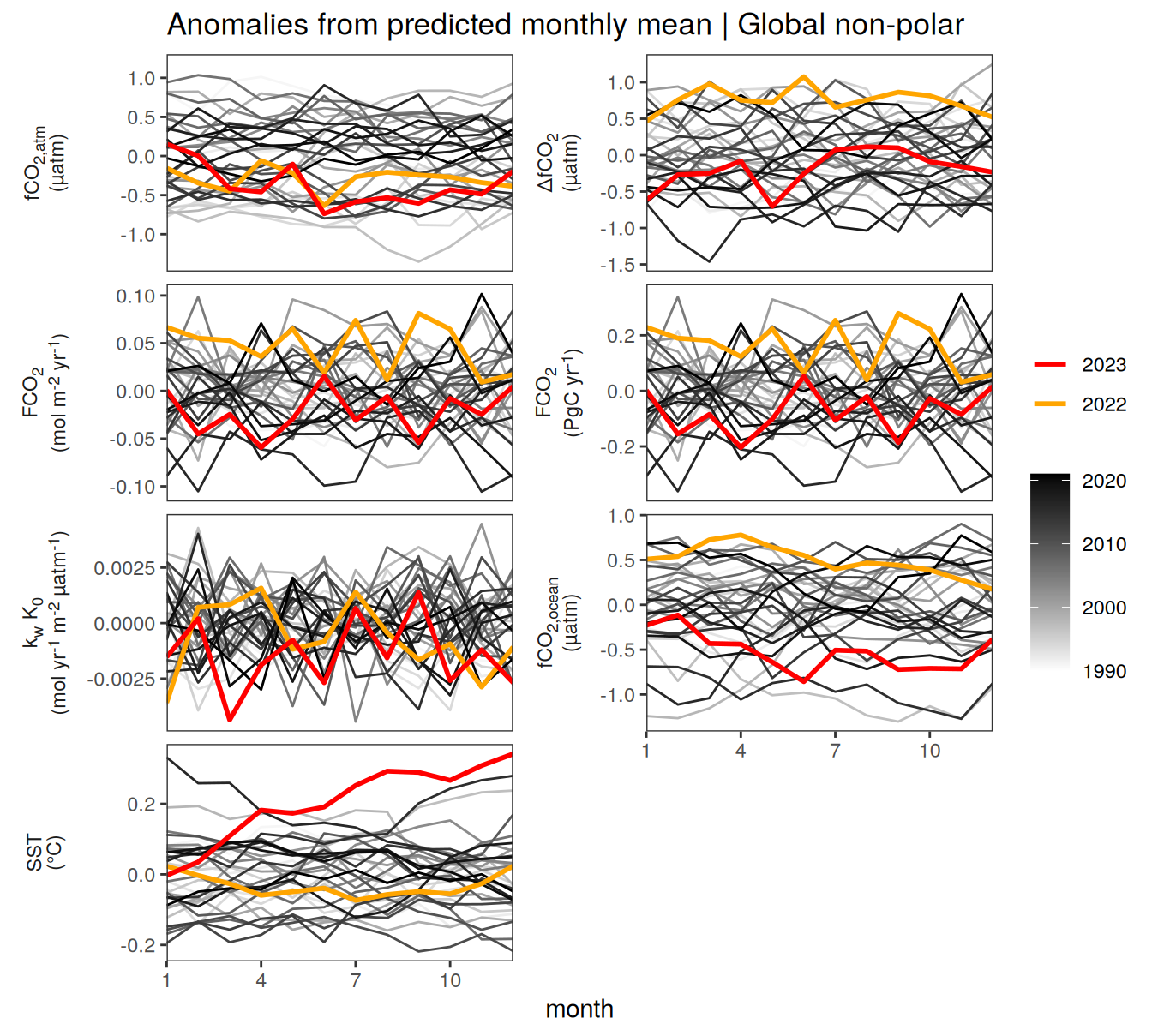
pco2_product_biome_monthly_detrended %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted monthly mean | Selected biomes") +
facet_grid(
name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)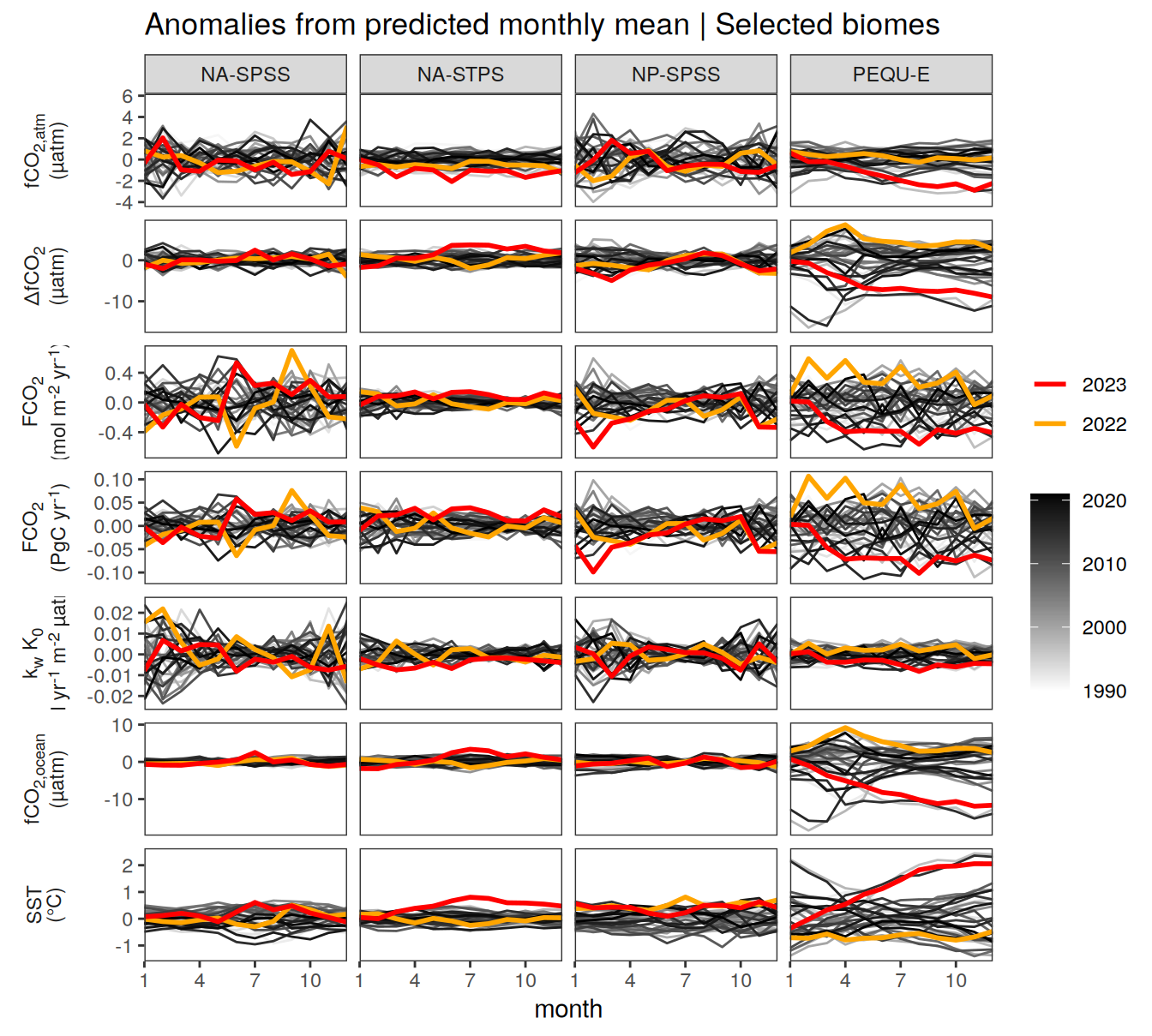
Key biomes
pco2_product_biome_monthly_detrended %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted monthly mean | Selected biomes") +
facet_grid(
name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)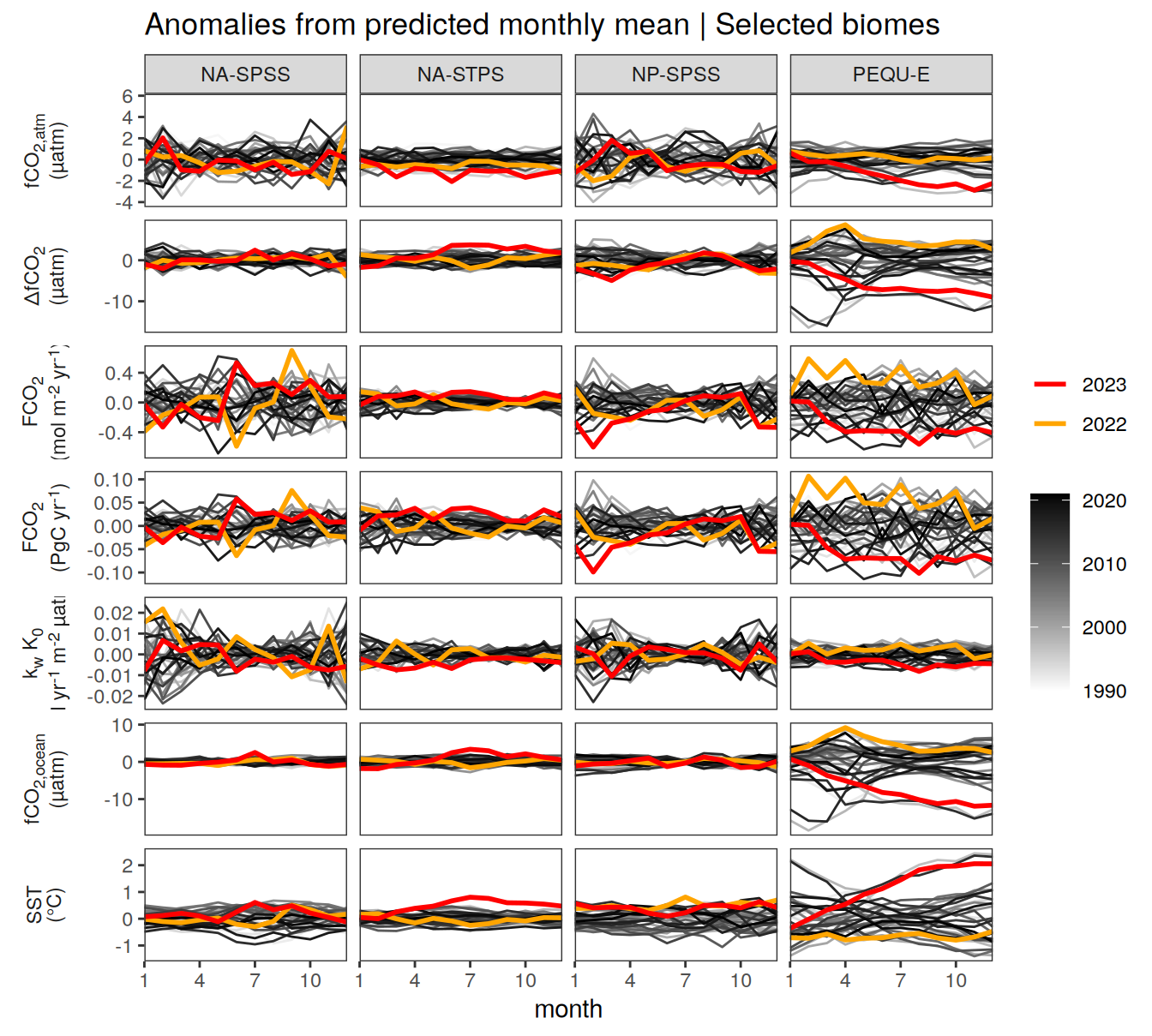
pco2_product_biome_monthly_detrended %>%
filter(biome %in% key_biomes) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(
data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1
) +
scale_color_manual(
values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = paste("Anomalies from predicted monthly mean |", .x$biome)) +
facet_wrap(
name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)
)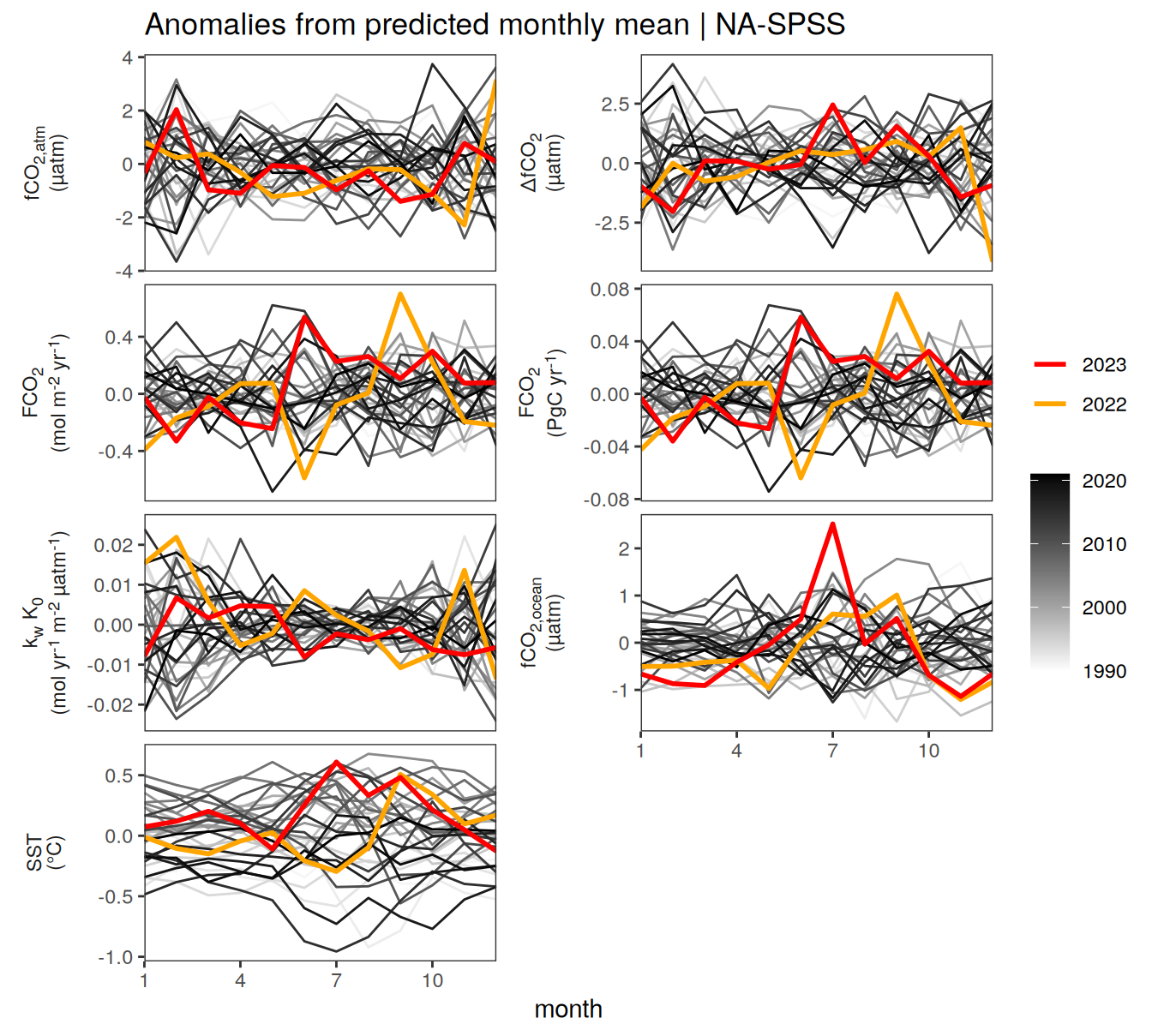
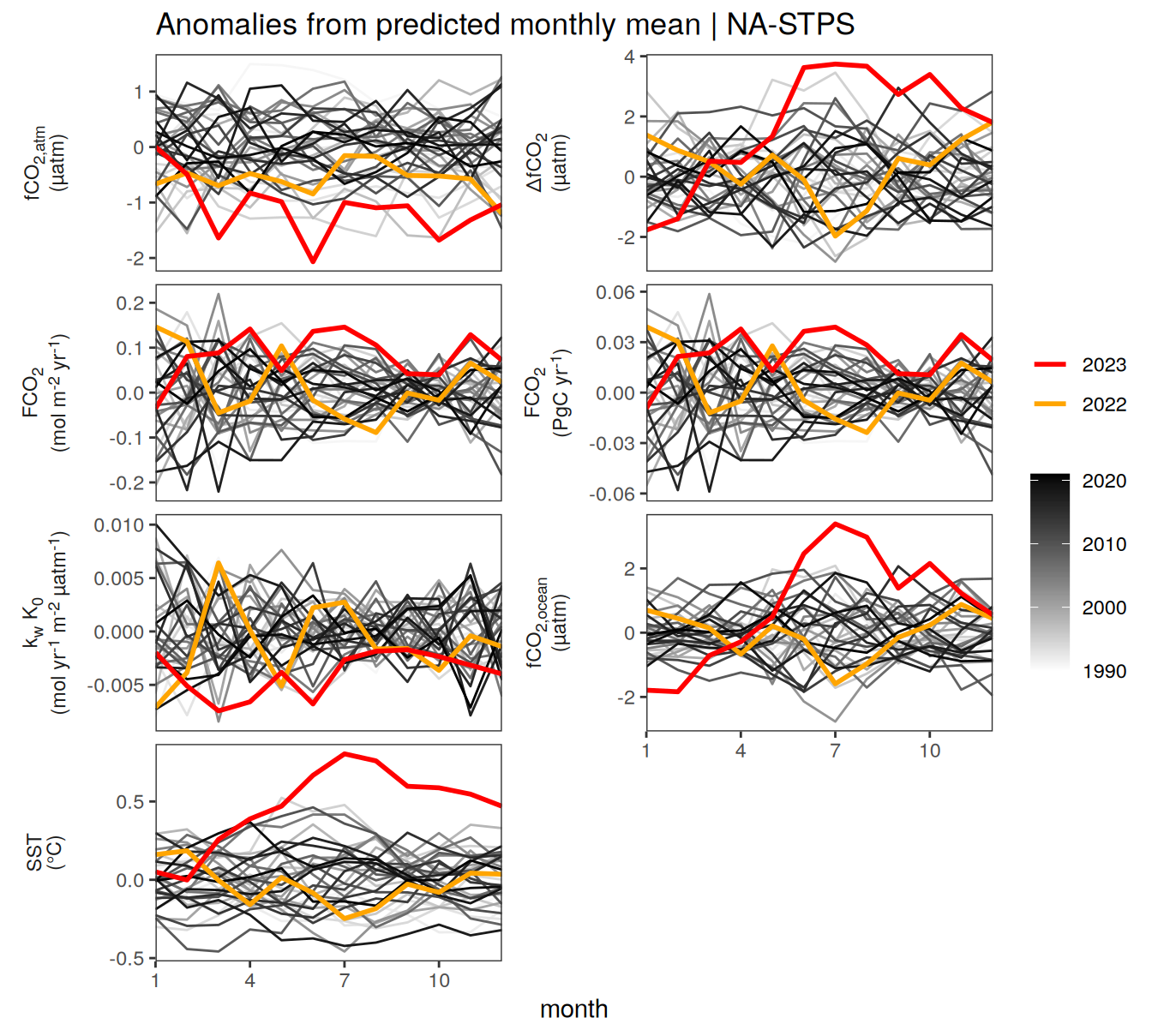
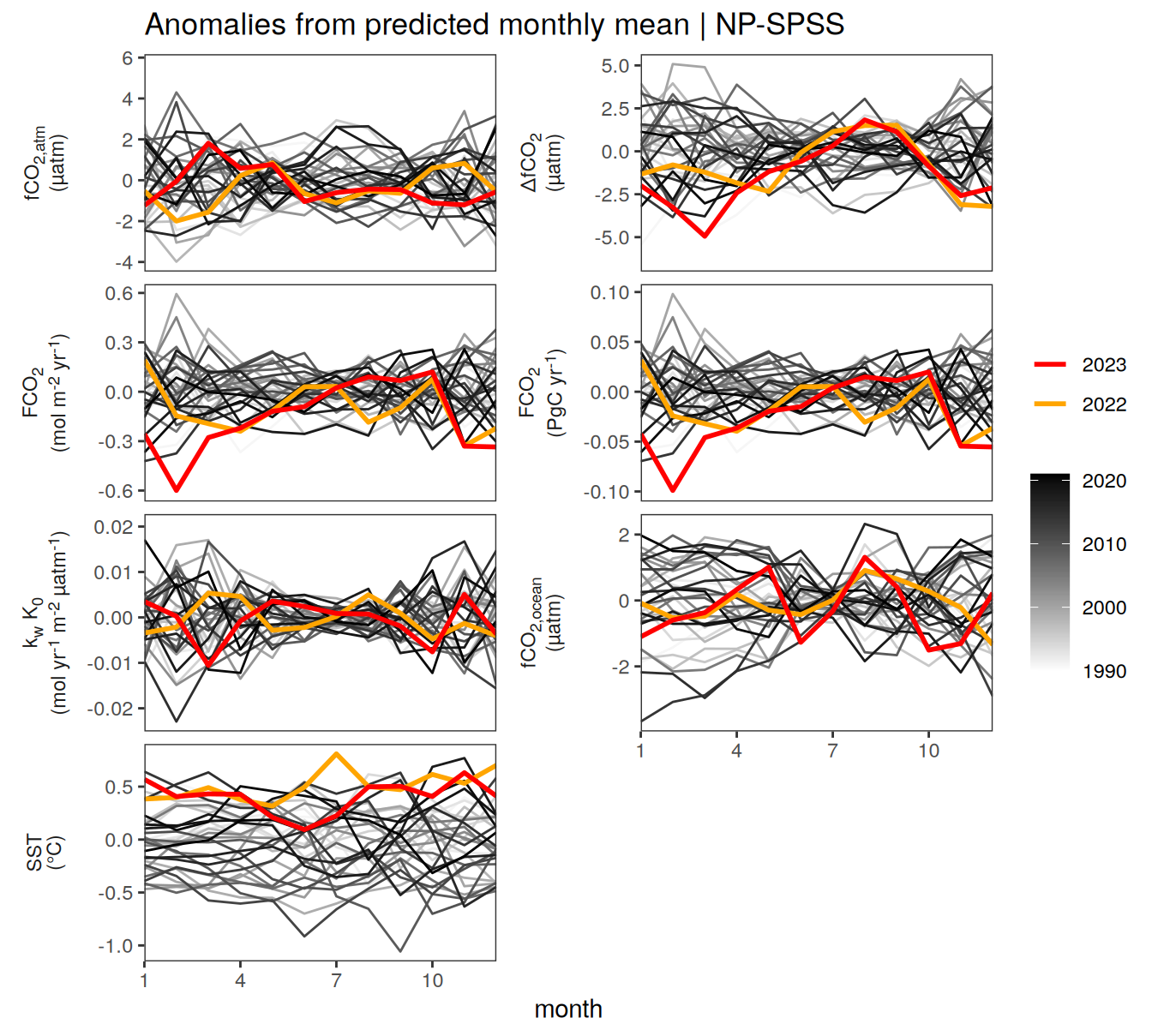
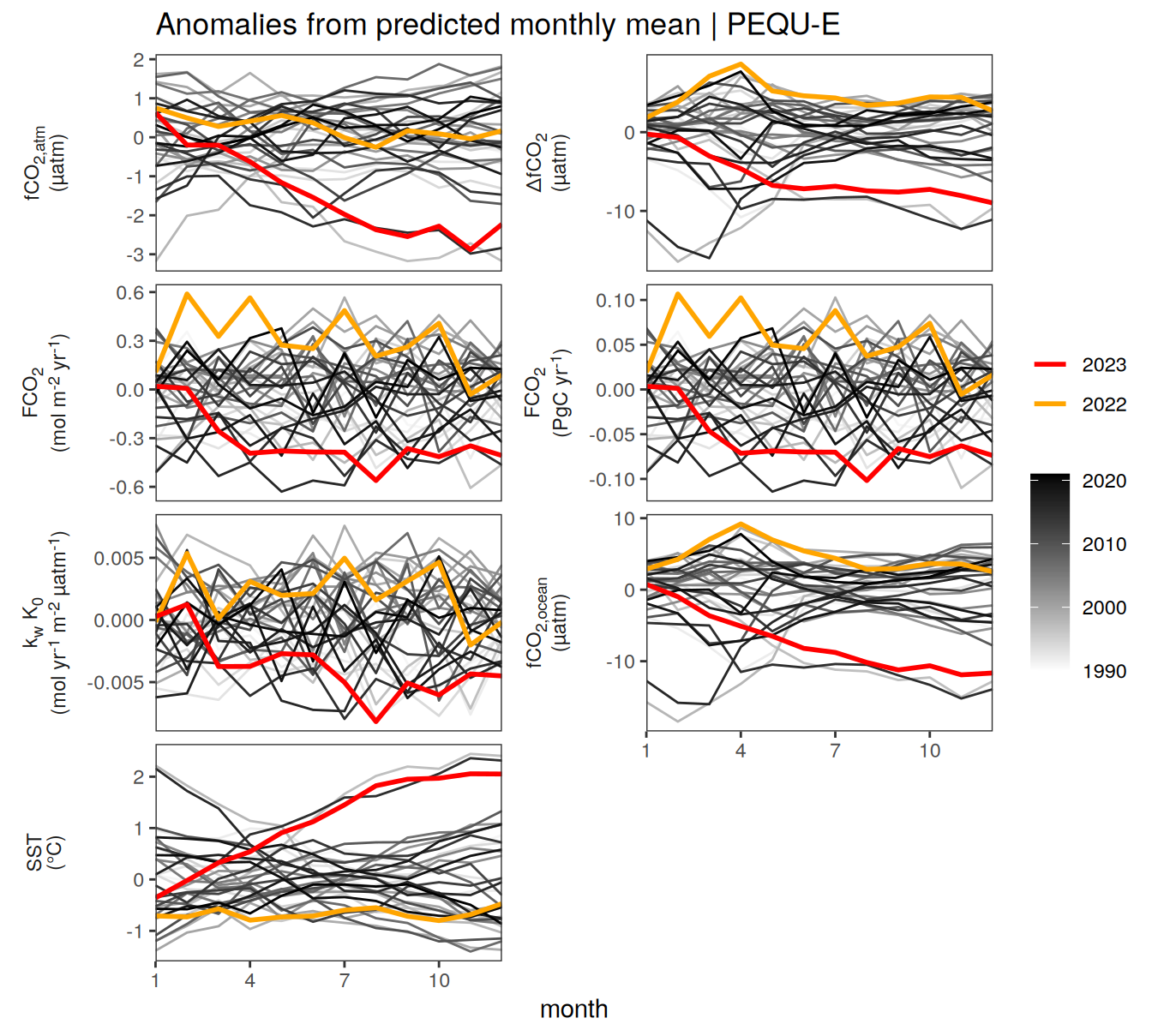
pco2_product_biome_monthly_detrended %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_monthly_detrended.csv"
)
)2023 anomaly correlation
The following plots aim to unravel the correlation between regionally integrated monthly flux anomalies and the corresponding anomalies of the means/integrals of each other variable.
Anomalies are first presented are first presented in absolute units. Due to the different flux magnitudes, we need to plot integrated fluxes separately for each region. Secondly, we normalize the monthly anomalies to the spread (expressed as standard deviation) of the residuals from the fit.
Annual anomalies
Absolute
pco2_product_biome_annual_anomaly %>%
filter(biome == "Global non-polar") %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(year, biome, fgco2_int)) %>%
ggplot(aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(fill = year),
shape = 21) +
geom_smooth(
data = . %>% filter(!between(year, 2023-1, 2023)),
method = "lm",
se = FALSE,
fullrange = TRUE,
aes(col = paste("Regression fit\nexcl.", 2023))
) +
scale_color_grey() +
scale_fill_grayC()+
new_scale_fill() +
geom_point(data = . %>% filter(between(year, 2023-1, 2023)),
aes(fill = as.factor(year)),
shape = 21, size = 2) +
scale_fill_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
labs(title = "Global non-polar integrated fluxes",
y = labels_breaks("fgco2_int")$i_legend_title) +
facet_wrap(
~ name,
scales = "free_x",
labeller = labeller(name = x_axis_labels),
strip.position = "bottom",
ncol = 2
) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.y = element_markdown(),
axis.title.x = element_blank(),
legend.title = element_blank()
)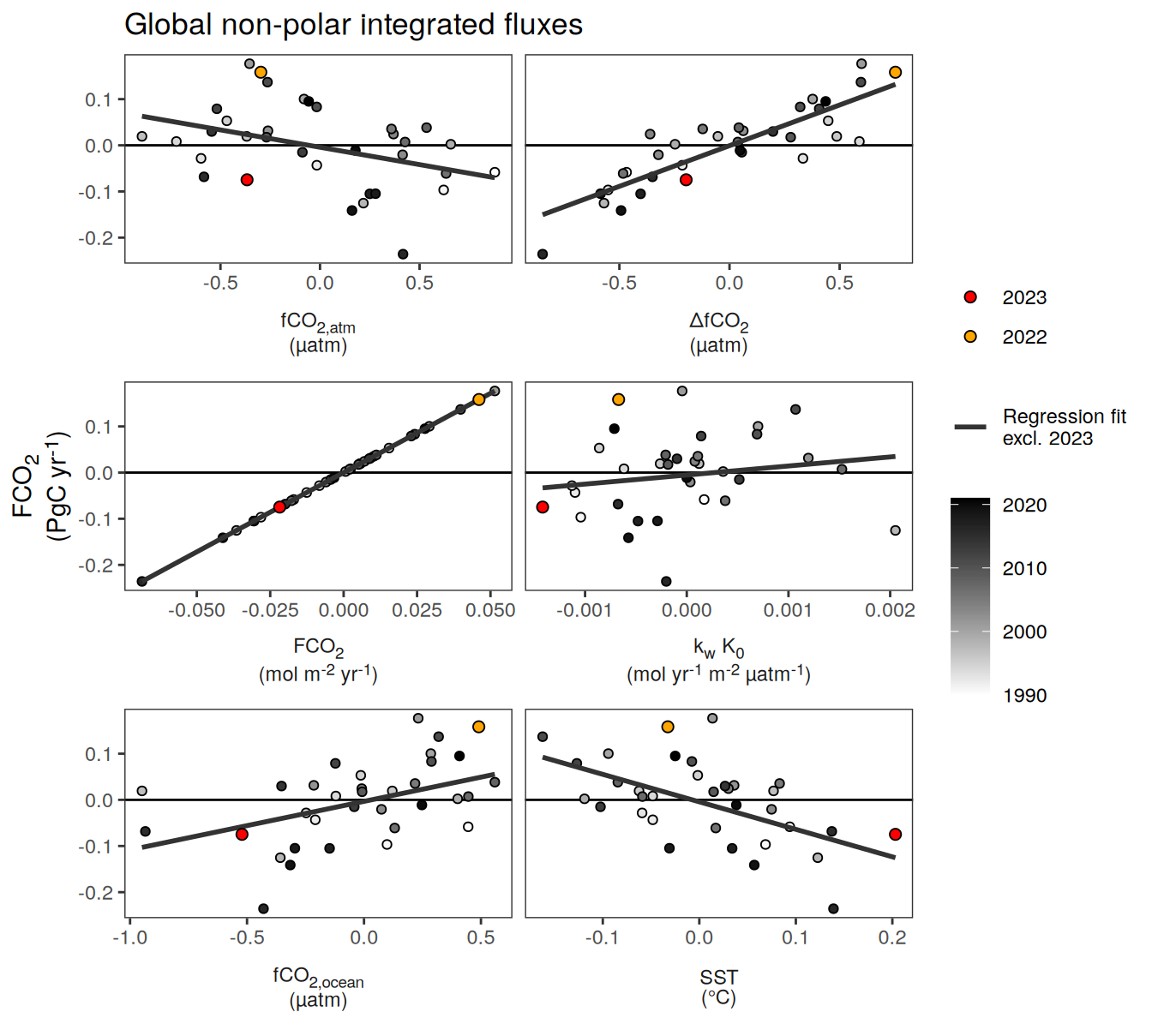
Monthly anomalies
Absolute
pco2_product_biome_monthly_detrended_anomaly <-
pco2_product_biome_monthly_detrended %>%
select(year, month, biome, name, resid) %>%
pivot_wider(names_from = name,
values_from = resid)
pco2_product_biome_monthly_detrended_anomaly %>%
filter(biome == "Global non-polar") %>%
pivot_longer(-c(year, month, biome, fgco2_int)) %>%
ggplot(aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3) +
scale_color_scico_d(palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)) +
scale_fill_scico_d(palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)) +
labs(title = "Global non-polar integrated fluxes",
y = labels_breaks("fgco2_int")$i_legend_title) +
facet_wrap(
~ name,
scales = "free_x",
labeller = labeller(name = x_axis_labels),
strip.position = "bottom",
ncol = 2
) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.y = element_markdown(),
axis.title.x = element_blank()
)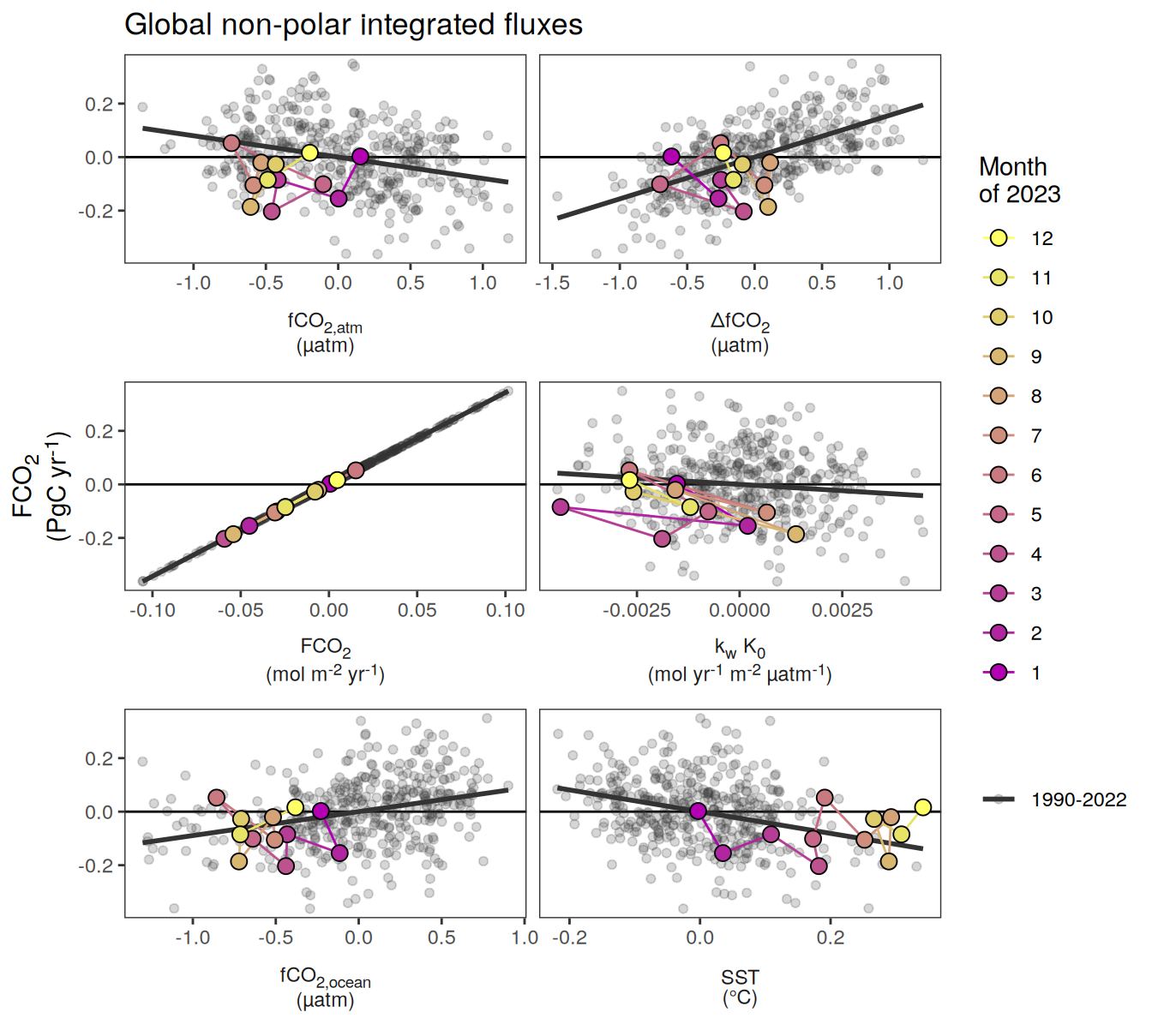
pco2_product_biome_monthly_detrended_anomaly %>%
filter(!(biome %in% c(key_biomes, "Global non-polar"))) %>%
pivot_longer(-c(year, month, biome, fgco2_int)) %>%
filter(name %in% c("temperature", "chl", "dfco2", "kw_sol")) %>%
group_split(name) %>%
head(1) %>%
map(
~ ggplot(data = .x,
aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2
) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(
data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3
) +
scale_color_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
scale_fill_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
facet_wrap( ~ biome, ncol = 3, scales = "free") +
labs(
title = "Biome integrated fluxes",
y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks(.x %>% distinct(name))$i_legend_title
) +
theme(axis.title.x = element_markdown(),
axis.title.y = element_markdown())
)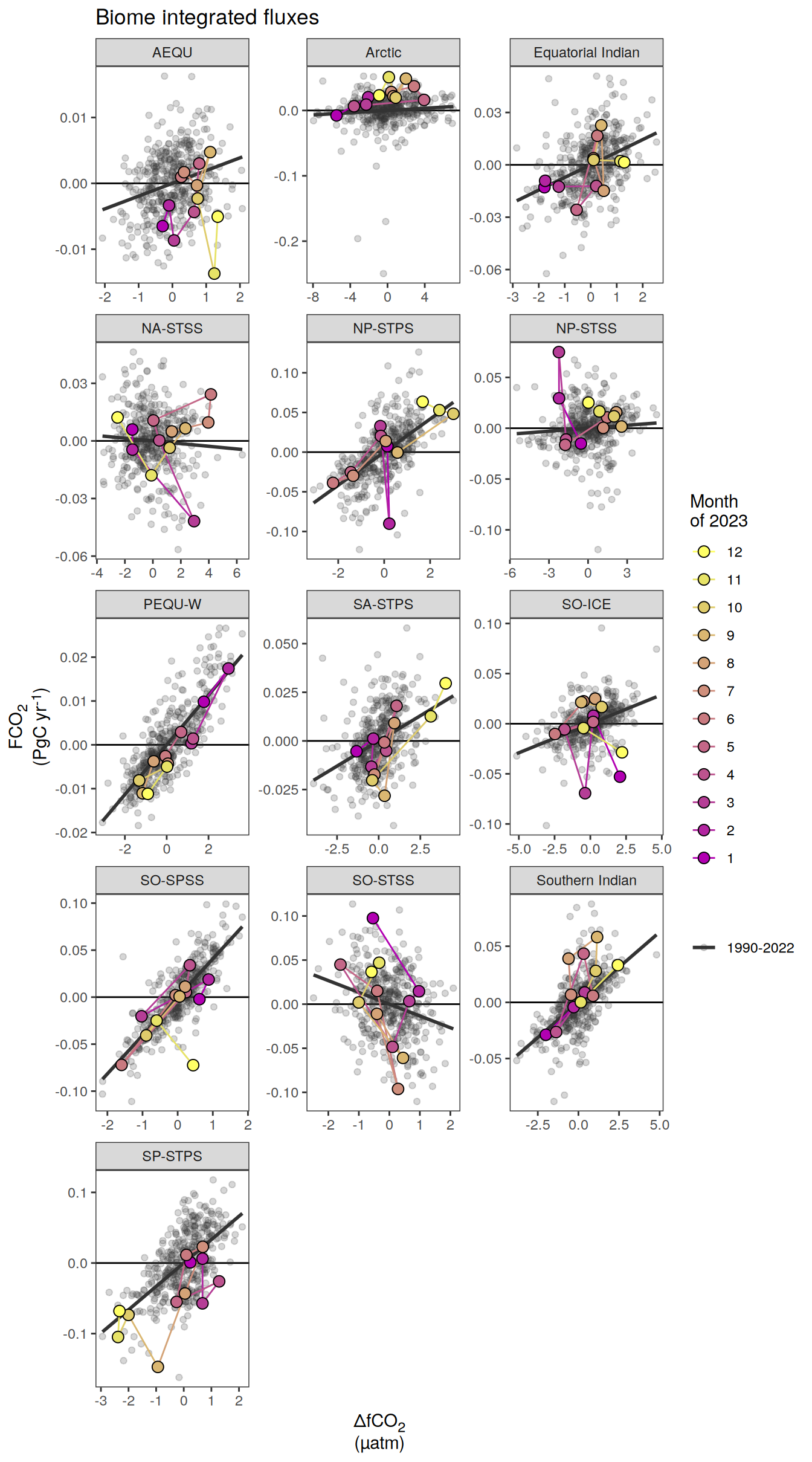
Relative to spread
pco2_product_biome_monthly_detrended_anomaly_spread <-
pco2_product_biome_monthly_detrended_anomaly %>%
pivot_longer(-c(month, biome, year)) %>%
filter(year != 2023) %>%
group_by(month, biome, name) %>%
summarise(spread = sd(value, na.rm = TRUE)) %>%
ungroup()
pco2_product_biome_monthly_detrended_anomaly_relative <-
full_join(
pco2_product_biome_monthly_detrended_anomaly_spread,
pco2_product_biome_monthly_detrended_anomaly %>%
pivot_longer(-c(month, biome, year))
)
pco2_product_biome_monthly_detrended_anomaly_relative <-
pco2_product_biome_monthly_detrended_anomaly_relative %>%
mutate(value = value / spread) %>%
select(-spread) %>%
pivot_wider() %>%
pivot_longer(-c(month, biome, year, fgco2_int))
pco2_product_biome_monthly_detrended_anomaly_relative %>%
filter(name %in% c("temperature", "chl", "dfco2", "kw_sol")) %>%
group_split(name) %>%
head(1) %>%
map(
~ ggplot(data = .x,
aes(value, fgco2_int)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 0) +
geom_point(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2
) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(
data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3
) +
scale_color_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
scale_fill_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
facet_wrap( ~ biome, ncol = 3) +
coord_fixed() +
labs(
title = "Biome integrated fluxes normalized to spread",
y = str_split_i(labels_breaks("fgco2_int")$i_legend_title, "<br>", i = 1),
x = str_split_i(labels_breaks(.x %>% distinct(name))$i_legend_title, "<br>", i = 1)
) +
theme(axis.title.x = element_markdown(),
axis.title.y = element_markdown())
)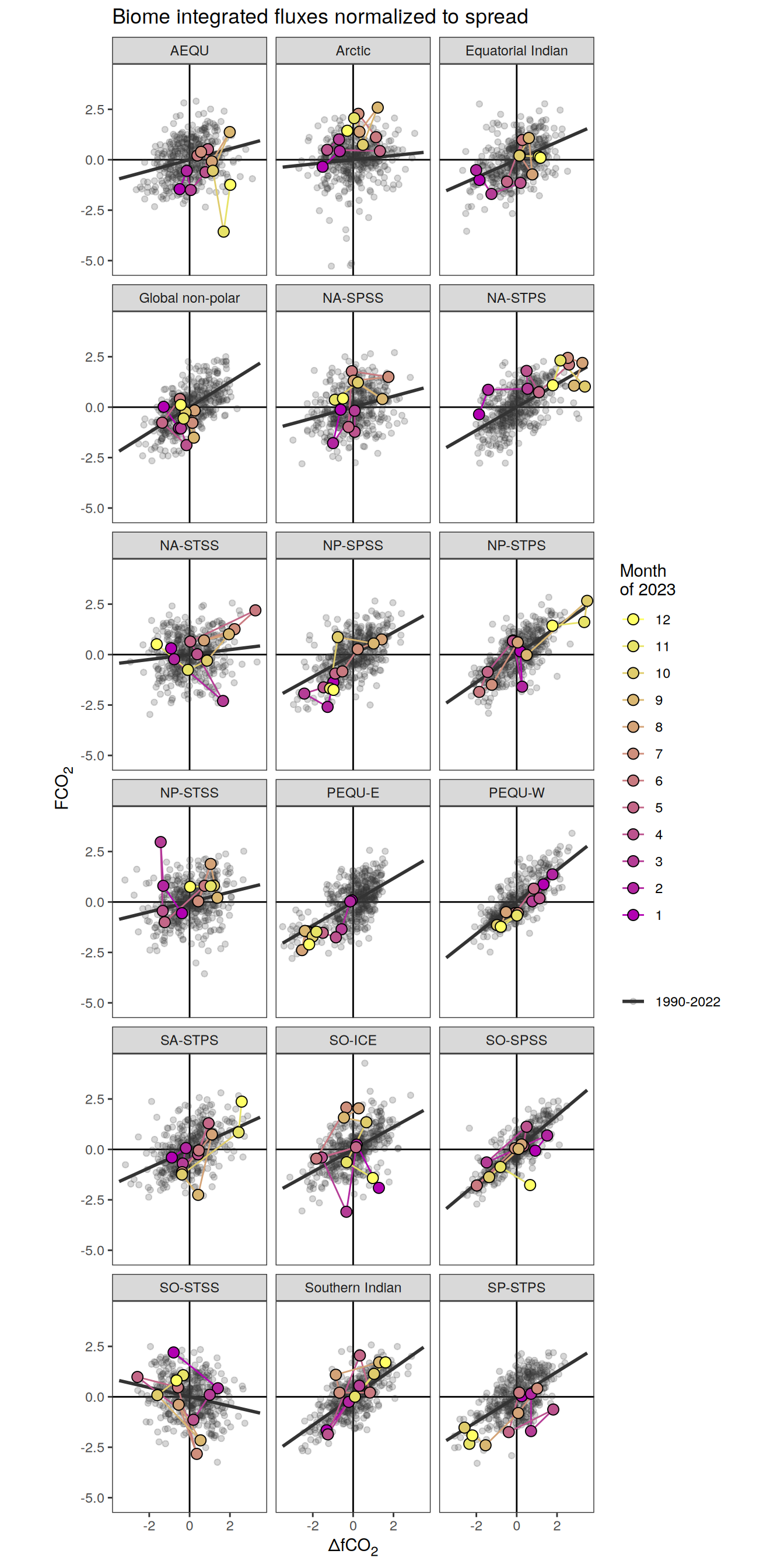
fCO2 decomposition
biome_mask <-
bind_rows(
biome_mask,
biome_mask %>%
filter(!str_detect(biome, "SO-SPSS|SO-ICE|Arctic")) %>%
mutate(biome = "Global non-polar")
)
pco2_product_biome_monthly_fCO2_decomposition <-
full_join(pco2_product_map_monthly_fCO2_decomposition,
biome_mask,
relationship = "many-to-many") %>%
group_by(year, month, biome, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup() %>%
drop_na()
pco2_product_biome_annual_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
group_by(year, biome, name) %>%
summarise(resid = mean(resid)) %>%
ungroup()
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
p_season(title = paste("Anomalies from predicted monthly mean"))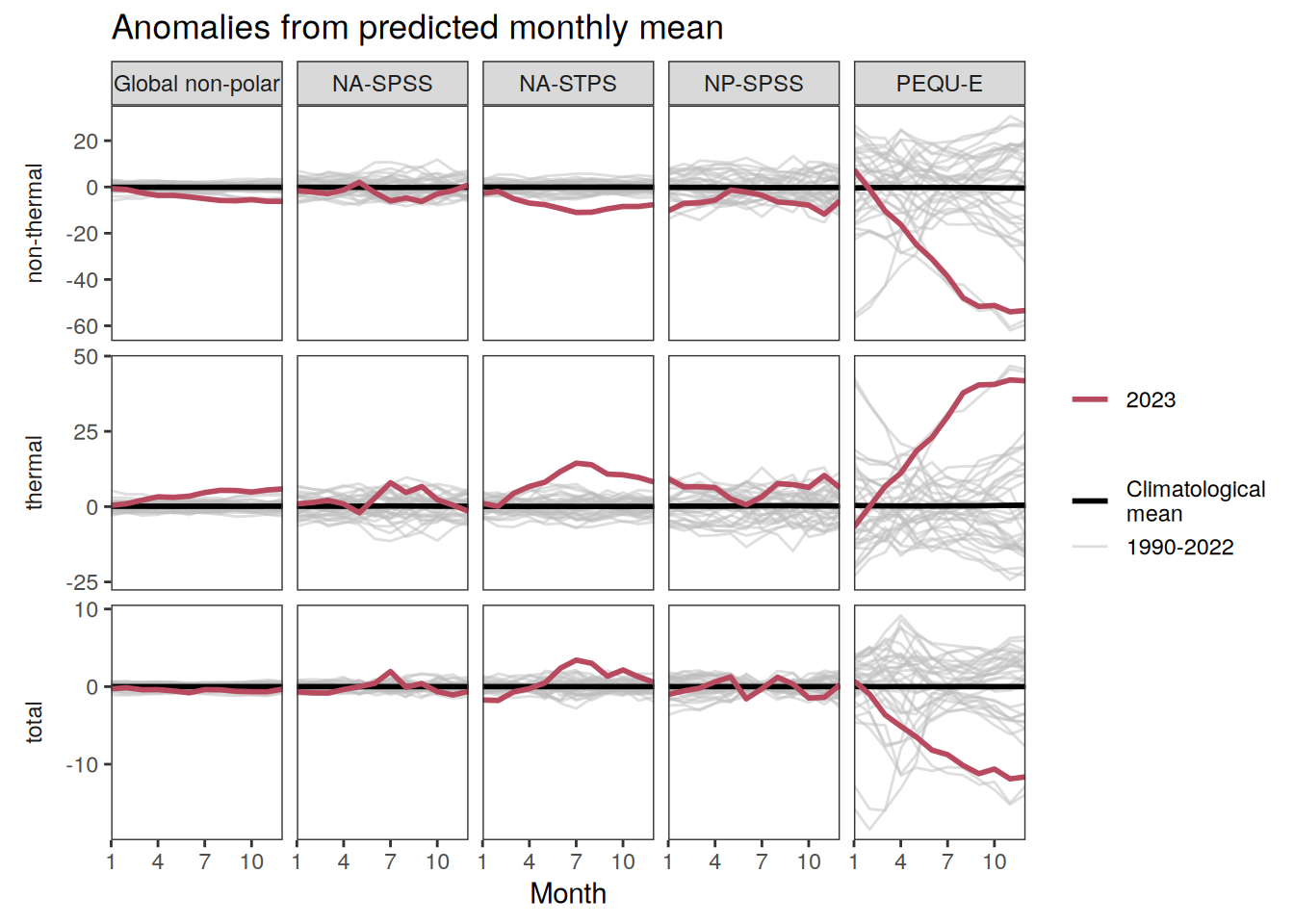
Flux attribution
Seasonal
pco2_product_biome_monthly_flux_attribution <-
full_join(pco2_product_map_monthly_flux_attribution,
biome_mask,
relationship = "many-to-many") %>%
group_by(year, month, biome, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup() %>%
drop_na()
pco2_product_biome_monthly_flux_attribution_total <-
full_join(pco2_product_map_monthly_anomaly %>%
filter(name == "fgco2") %>%
mutate(name = "resid_fgco2"),
biome_mask,
relationship = "many-to-many") %>%
group_by(year, month, biome, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup() %>%
drop_na()
pco2_product_biome_monthly_flux_attribution <-
bind_rows(
pco2_product_biome_monthly_flux_attribution,
pco2_product_biome_monthly_flux_attribution_total
)
pco2_product_biome_annual_flux_attribution <-
pco2_product_biome_monthly_flux_attribution %>%
group_by(year, biome, name) %>%
summarise(resid = mean(resid)) %>%
ungroup()
pco2_product_biome_monthly_flux_attribution %>%
filter(year == 2023,
biome %in% c("Global non-polar", key_biomes)) %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_path(
aes(month, resid)
) +
geom_point(
aes(month, resid),
shape = 21,
alpha = 0.5,
col = "grey30"
) +
scale_y_continuous(breaks = seq(-10,10,0.2)) +
scale_x_continuous(position = "top", breaks = seq(1,12,3)) +
labs(y = labels_breaks(unique("fgco2"))$i_legend_title) +
facet_grid(biome ~ name,
labeller = labeller(name = x_axis_labels),
scales = "free_y",
space = "free_y",
switch = "x") +
theme(
legend.title = element_blank(),
axis.title.y = element_markdown(),
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
legend.position = "top"
)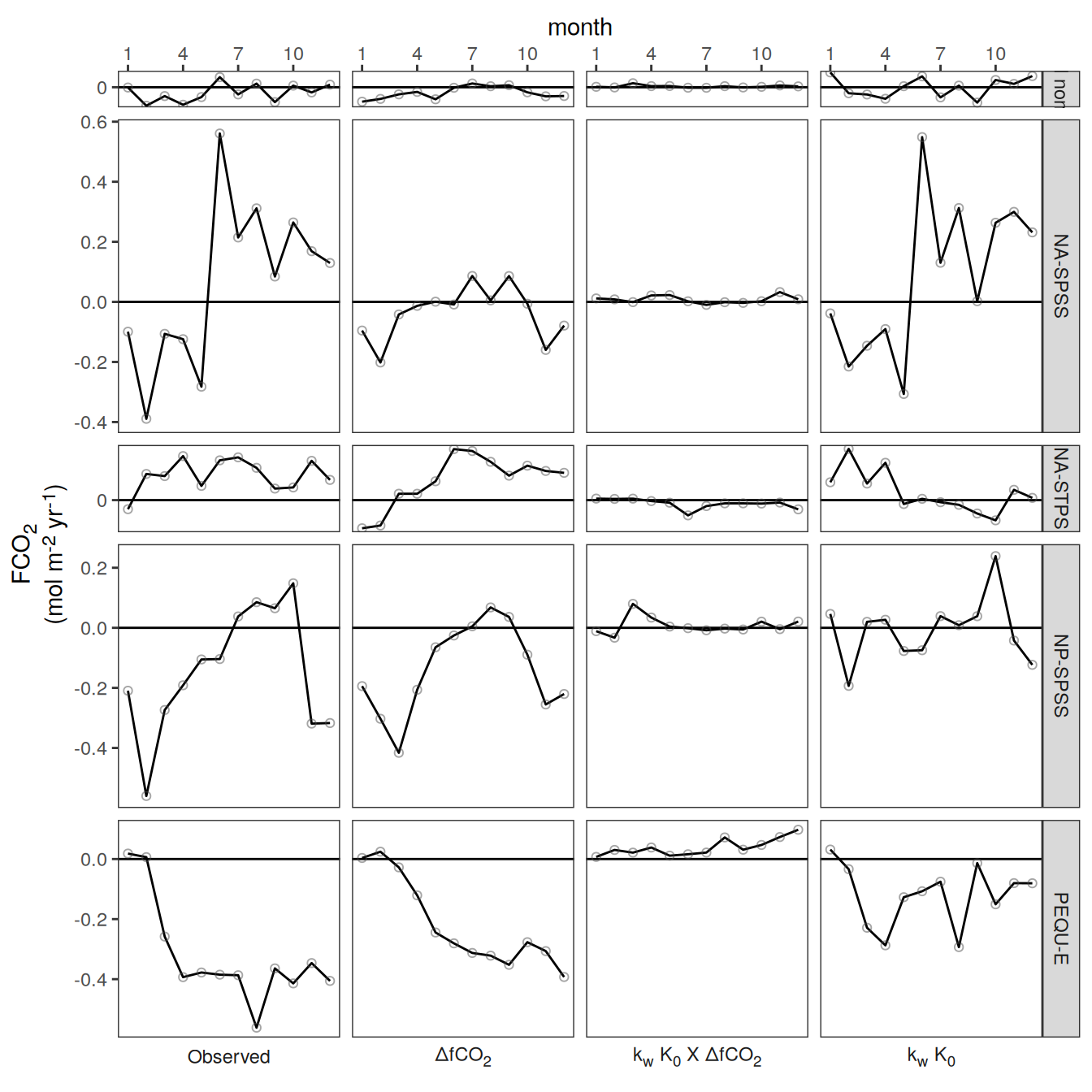
pco2_product_biome_monthly_flux_attribution %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
p_season(title = paste("Anomalies from predicted monthly mean"))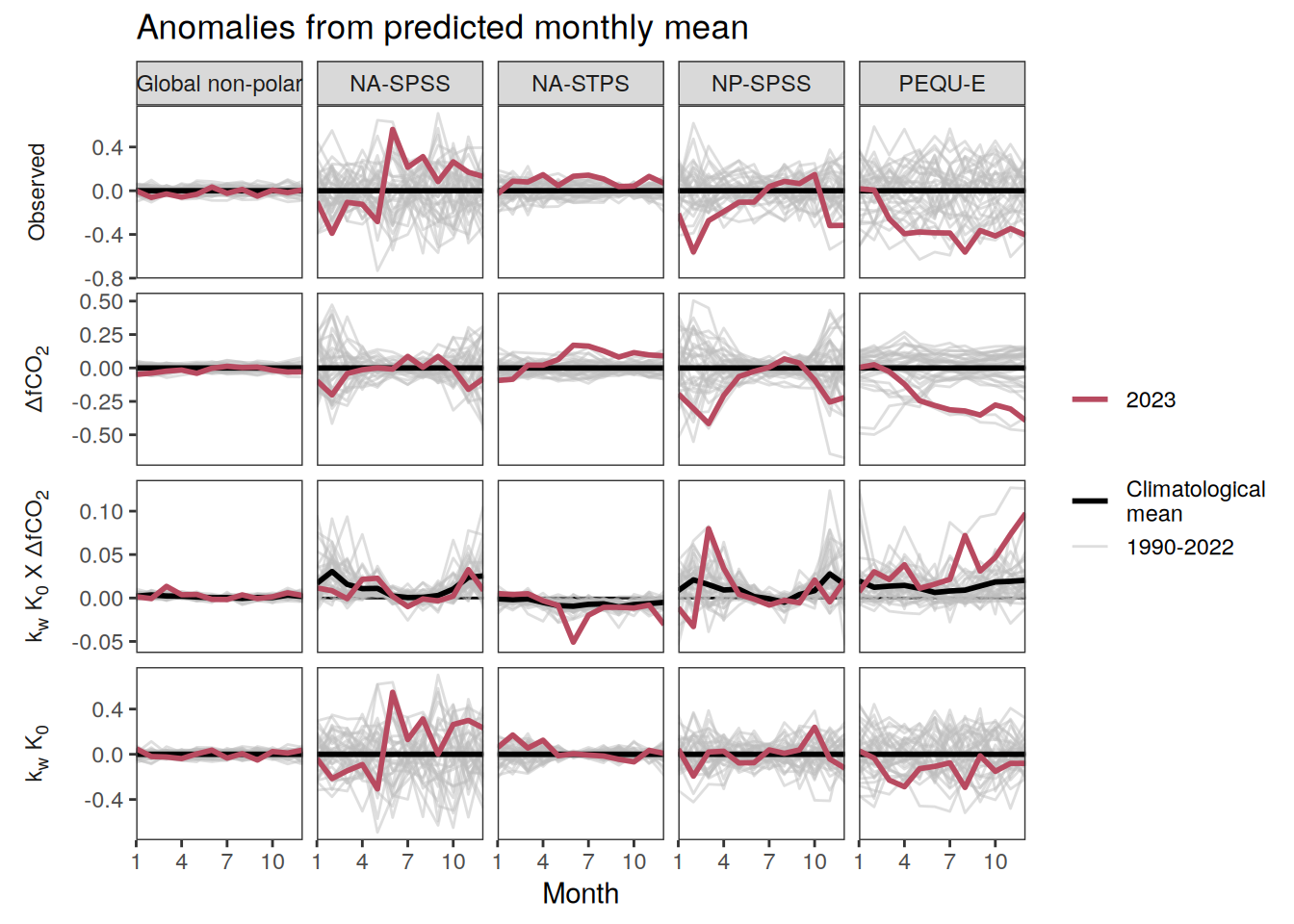
Annual
pco2_product_biome_annual_flux_attribution <-
full_join(
pco2_product_biome_annual_flux_attribution %>%
filter(year == 2023) %>%
select(-year),
pco2_product_biome_annual_flux_attribution %>%
filter(year != 2023) %>%
group_by(biome, name) %>%
summarise(resid_mean = mean(abs(resid))) %>%
ungroup()
)
# pco2_product_biome_annual_flux_attribution %>%
# filter(biome %in% c("Global non-polar", key_biomes)) %>%
# mutate(product == "pco2 product") %>%
# group_split(product) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x) +
# geom_col(aes("x", resid),
# position = "dodge2") +
# geom_col(
# aes(
# "x",
# resid_mean * sign(resid),
# col = paste0("Mean\nexcl.",2023)
# ),
# position = "dodge2",
# fill = "transparent"
# ) +
# labs(y = labels_breaks(unique("fgco2"))$i_legend_title,
# title = .x$biome) +
# facet_grid(
# biome~name,
# labeller = labeller(name = x_axis_labels),
# switch = "x"
# ) +
# scale_color_grey() +
# theme(
# legend.title = element_blank(),
# axis.text.x = element_blank(),
# axis.ticks.x = element_blank(),
# axis.title.x = element_blank(),
# axis.title.y = element_markdown(),
# strip.text.x.bottom = element_markdown(),
# strip.placement = "outside",
# strip.background.x = element_blank(),
# legend.position = "top"
# )
# )pco2_product_biome_annual_flux_attribution %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_annual_flux_attribution.csv"
)
)
pco2_product_biome_monthly_flux_attribution %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_monthly_flux_attribution.csv"
)
)
pco2_product_biome_annual_fCO2_decomposition %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_annual_fCO2_decomposition.csv"
)
)
pco2_product_biome_monthly_fCO2_decomposition %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_biome_monthly_fCO2_decomposition.csv"
)
)
rm(
pco2_product_biome_annual_flux_attribution,
pco2_product_biome_monthly_flux_attribution,
pco2_product_biome_annual_fCO2_decomposition,
pco2_product_biome_monthly_fCO2_decomposition
)
gc()Zonal mean sections
The following analysis is available for GOBMs only.
Annual means
2023 anomaly
pco2_product_zonal_mean_annual <- pco2_product_zonal_mean %>%
pivot_longer(-c(region, depth, lat, time, year, month)) %>%
group_by(region, lat, depth, year, name) %>%
summarise(value = mean(value)) %>%
ungroup() %>%
drop_na() %>%
mutate(region = str_to_title(region))
pco2_product_zonal_mean_annual_anomaly <-
pco2_product_zonal_mean_annual %>%
anomaly_determination(region, lat, depth)
pco2_product_zonal_mean_annual_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(3) %>%
map(
~ ggplot(data = .x) +
geom_contour_filled(aes(lat, depth, z = resid)) +
scale_fill_discrete_divergingx(name = labels_breaks(.x %>% distinct(name))$i_legend_title) +
guides(fill = guide_colorsteps(
barheight = unit(8, "cm"),
show.limits = TRUE
)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50,100,200,400)) +
scale_x_continuous(breaks = seq(-100, 100, 20)) +
coord_cartesian(expand = 0) +
facet_wrap( ~ region, ncol = 1) +
labs(y = "Depth (m)") +
theme(legend.title = element_markdown())
)pco2_product_zonal_mean_annual_anomaly %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_zonal_mean_sections_annual.csv"
)
)Biome profiles
The following analysis is available for GOBMs only.
Annual means
2023 anomaly
pco2_product_profiles_annual <- pco2_product_profiles %>%
pivot_longer(-c(biome, depth, time, year, month)) %>%
group_by(biome, depth, year, name) %>%
summarise(value = mean(value)) %>%
ungroup() %>%
drop_na()
pco2_product_profiles_annual_anomaly <-
pco2_product_profiles_annual %>%
anomaly_determination(biome, depth)
pco2_product_profiles_annual_anomaly %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_path(aes(resid, depth, group = year), col = "grey30", alpha = 0.5) +
geom_path(data = .x %>% filter(year == 2023),
aes(resid, depth, col = as.factor(year)),
linewidth = 1) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50,100,200,400)) +
facet_wrap( ~ biome) +
labs(y = "Depth (m)",
x = labels_breaks(.x %>% distinct(name))$i_legend_title) +
theme(legend.title = element_blank(),
axis.title.x = element_markdown())
)pco2_product_profiles_annual_anomaly %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_profiles_annual.csv"
)
)Monthly means
2023 anomaly
pco2_product_profiles_monthly <- pco2_product_profiles %>%
pivot_longer(-c(biome, depth, time, year, month)) %>%
group_by(biome, depth, year, month, name) %>%
summarise(value = mean(value)) %>%
ungroup() %>%
drop_na()
pco2_product_profiles_monthly_anomaly <-
pco2_product_profiles_monthly %>%
anomaly_determination(biome, depth, month)
pco2_product_profiles_monthly_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_vline(xintercept = 0) +
geom_path(aes(resid, depth, col = as.factor(month)),
linewidth = 1) +
scale_color_scico_d(palette = "hawaii") +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50, 100, 200, 400)) +
facet_wrap(~ biome,
scales = "free_x") +
labs(y = "Depth (m)",
x = labels_breaks(.x %>% distinct(name))$i_legend_title) +
theme(legend.title = element_blank(),
axis.title.x = element_markdown())
)pco2_product_profiles_monthly_anomaly %>%
write_csv(
paste0(
"../data/",
"NIES-ML3_GCB",
"_",
"2023",
"_profiles_monthly.csv"
)
)
sessionInfo()R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: openSUSE Leap 15.6
Matrix products: default
BLAS/LAPACK: /usr/local/OpenBLAS-0.3.28/lib/libopenblas_haswellp-r0.3.28.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Zurich
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggh4x_0.3.0 scales_1.3.0 cmocean_0.3-2
[4] ggtext_0.1.2 broom_1.0.7 khroma_1.14.0
[7] ggnewscale_0.5.0 seacarb_3.3.3 SolveSAPHE_2.1.0
[10] oce_1.8-3 gsw_1.2-0 stars_0.6-7
[13] abind_1.4-8 terra_1.8-5 sf_1.0-19
[16] rnaturalearth_1.0.1 geomtextpath_0.1.4 colorspace_2.1-1
[19] marelac_2.1.11 shape_1.4.6.1 ggforce_0.4.2
[22] metR_0.16.0 scico_1.5.0 patchwork_1.3.0
[25] collapse_2.0.18 lubridate_1.9.3 forcats_1.0.0
[28] stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[31] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[34] ggplot2_3.5.1 tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] DBI_1.2.3 rlang_1.1.4 magrittr_2.0.3
[4] git2r_0.35.0 e1071_1.7-16 compiler_4.4.2
[7] mgcv_1.9-1 getPass_0.2-4 systemfonts_1.1.0
[10] callr_3.7.6 vctrs_0.6.5 pkgconfig_2.0.3
[13] crayon_1.5.3 fastmap_1.2.0 backports_1.5.0
[16] ncmeta_0.4.0 labeling_0.4.3 utf8_1.2.4
[19] promises_1.3.2 rmarkdown_2.29 markdown_1.13
[22] tzdb_0.4.0 ps_1.8.1 bit_4.5.0
[25] xfun_0.49 cachem_1.1.0 jsonlite_1.8.9
[28] later_1.4.1 tweenr_2.0.3 parallel_4.4.2
[31] R6_2.5.1 RColorBrewer_1.1-3 bslib_0.8.0
[34] stringi_1.8.4 jquerylib_0.1.4 Rcpp_1.0.13-1
[37] knitr_1.49 Matrix_1.7-1 splines_4.4.2
[40] httpuv_1.6.15 timechange_0.3.0 tidyselect_1.2.1
[43] RNetCDF_2.9-2 rstudioapi_0.17.1 yaml_2.3.10
[46] codetools_0.2-20 processx_3.8.4 lattice_0.22-6
[49] withr_3.0.2 evaluate_1.0.1 rnaturalearthdata_1.0.0
[52] units_0.8-5 proxy_0.4-27 polyclip_1.10-7
[55] xml2_1.3.6 pillar_1.9.0 whisker_0.4.1
[58] CFtime_1.4.1 KernSmooth_2.23-24 checkmate_2.3.2
[61] generics_0.1.3 vroom_1.6.5 rprojroot_2.0.4
[64] hms_1.1.3 commonmark_1.9.2 munsell_0.5.1
[67] class_7.3-22 glue_1.8.0 tools_4.4.2
[70] data.table_1.16.2 fs_1.6.5 grid_4.4.2
[73] nlme_3.1-166 cli_3.6.3 textshaping_0.4.0
[76] fansi_1.0.6 viridisLite_0.4.2 gtable_0.3.6
[79] sass_0.4.9 digest_0.6.37 classInt_0.4-10
[82] farver_2.1.2 memoise_2.0.1 htmltools_0.5.8.1
[85] lifecycle_1.0.4 httr_1.4.7 here_1.0.1
[88] gridtext_0.1.5 bit64_4.5.2 MASS_7.3-61