pCO2 product synopsis 2023
Jens Daniel Müller
12 March, 2025
Last updated: 2025-03-12
Checks: 7 0
Knit directory:
heatwave_co2_flux_2023/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240307) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3a5d484. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data
Ignored: output/
Unstaged changes:
Modified: analysis/child/pCO2_product_synopsis.Rmd
Modified: code/Workflowr_project_managment.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/pco2_product_synopsis_2023.Rmd) and HTML
(docs/pco2_product_synopsis_2023.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 3a5d484 | jens-daniel-mueller | 2025-03-11 | Build site. |
| html | b031240 | jens-daniel-mueller | 2025-03-11 | Build site. |
| html | 17f843d | jens-daniel-mueller | 2025-03-10 | Build site. |
| html | b9e8f8c | jens-daniel-mueller | 2025-03-10 | Build site. |
| html | 60900ed | jens-daniel-mueller | 2025-03-07 | Build site. |
| html | 03a727b | jens-daniel-mueller | 2025-03-07 | Build site. |
| html | dd25c0c | jens-daniel-mueller | 2025-03-06 | Build site. |
| html | f2beca5 | jens-daniel-mueller | 2025-03-04 | Build site. |
| html | 89d6687 | jens-daniel-mueller | 2025-03-02 | Build site. |
| html | 528798e | jens-daniel-mueller | 2025-02-27 | Build site. |
| Rmd | 6fd2de0 | jens-daniel-mueller | 2025-02-27 | testrun external file definition |
| html | e717448 | jens-daniel-mueller | 2025-02-26 | Build site. |
| html | 709a2cf | jens-daniel-mueller | 2025-02-26 | Build site. |
| html | 4d090e2 | jens-daniel-mueller | 2025-02-25 | Build site. |
| html | 3548f48 | jens-daniel-mueller | 2025-02-25 | Build site. |
| html | 1d3f08c | jens-daniel-mueller | 2025-02-24 | Build site. |
| html | 4d537ca | jens-daniel-mueller | 2025-02-23 | Build site. |
| html | 945d8f7 | jens-daniel-mueller | 2025-02-23 | Build site. |
| html | d3235fa | jens-daniel-mueller | 2024-09-20 | Build site. |
| html | 713d232 | jens-daniel-mueller | 2024-09-12 | Build site. |
| html | 878c674 | jens-daniel-mueller | 2024-09-10 | Build site. |
| html | 4acb1fc | jens-daniel-mueller | 2024-09-05 | Build site. |
| html | 3bb8433 | jens-daniel-mueller | 2024-09-03 | Build site. |
| html | c50054d | jens-daniel-mueller | 2024-08-29 | Build site. |
| html | aecb187 | jens-daniel-mueller | 2024-08-28 | Build site. |
| html | 0f5b472 | jens-daniel-mueller | 2024-08-27 | Build site. |
| html | 08ca8c7 | jens-daniel-mueller | 2024-08-27 | Build site. |
| html | d82bd91 | jens-daniel-mueller | 2024-08-27 | Build site. |
| html | 681629f | jens-daniel-mueller | 2024-08-26 | Build site. |
| html | 6a96e1f | jens-daniel-mueller | 2024-08-26 | Build site. |
| html | c62d92d | jens-daniel-mueller | 2024-08-23 | Build site. |
| html | 2f165ec | jens-daniel-mueller | 2024-07-23 | Build site. |
| html | 2a28b07 | jens-daniel-mueller | 2024-07-22 | Build site. |
| html | 4ef742b | jens-daniel-mueller | 2024-07-20 | Build site. |
| html | a58162a | jens-daniel-mueller | 2024-07-11 | Build site. |
| html | 4a437fb | jens-daniel-mueller | 2024-07-09 | Build site. |
| html | 4897f6e | jens-daniel-mueller | 2024-07-08 | Build site. |
| html | ba4aaac | jens-daniel-mueller | 2024-07-08 | Build site. |
| html | 67956dd | jens-daniel-mueller | 2024-07-08 | Build site. |
| html | b7806ad | jens-daniel-mueller | 2024-07-02 | Build site. |
| html | dd97823 | jens-daniel-mueller | 2024-06-28 | Build site. |
| html | c6f967e | jens-daniel-mueller | 2024-06-28 | Build site. |
| html | b18b0e5 | jens-daniel-mueller | 2024-06-28 | Build site. |
| html | 197dac4 | jens-daniel-mueller | 2024-06-27 | Build site. |
| html | 9589349 | jens-daniel-mueller | 2024-06-27 | Build site. |
| html | 8cdfed7 | jens-daniel-mueller | 2024-06-21 | Build site. |
| html | aeca619 | jens-daniel-mueller | 2024-06-19 | Build site. |
| html | 478e699 | jens-daniel-mueller | 2024-06-14 | Build site. |
| html | d2a80a9 | jens-daniel-mueller | 2024-06-14 | Build site. |
| html | f03b1d8 | jens-daniel-mueller | 2024-06-12 | Build site. |
| html | d46002d | jens-daniel-mueller | 2024-06-12 | manual commit |
| html | 34b4fe2 | jens-daniel-mueller | 2024-06-12 | Build site. |
| html | 5261667 | jens-daniel-mueller | 2024-06-11 | manual commit |
| html | 2b34bf8 | jens-daniel-mueller | 2024-06-11 | manual commit |
| html | 6954c65 | jens-daniel-mueller | 2024-06-06 | Build site. |
| html | 54af933 | jens-daniel-mueller | 2024-06-03 | Build site. |
| html | 2454031 | jens-daniel-mueller | 2024-06-01 | Build site. |
| html | e83b65a | jens-daniel-mueller | 2024-05-31 | Build site. |
| html | 7919445 | jens-daniel-mueller | 2024-05-31 | Build site. |
| html | 6fc213f | jens-daniel-mueller | 2024-05-31 | Build site. |
| html | 7c448f7 | jens-daniel-mueller | 2024-05-31 | Build site. |
| html | bf01e6c | jens-daniel-mueller | 2024-05-31 | Build site. |
| html | 4e9d7c0 | jens-daniel-mueller | 2024-05-30 | Build site. |
| html | fc1b92d | jens-daniel-mueller | 2024-05-30 | Build site. |
| html | 4d3ccb2 | jens-daniel-mueller | 2024-05-29 | Build site. |
| html | 7ad8576 | jens-daniel-mueller | 2024-05-29 | Build site. |
| html | 0493049 | jens-daniel-mueller | 2024-05-29 | Build site. |
| html | acaac5f | jens-daniel-mueller | 2024-05-28 | Build site. |
| html | b99b329 | jens-daniel-mueller | 2024-05-28 | Build site. |
| html | b754e95 | jens-daniel-mueller | 2024-05-28 | Build site. |
| html | fbba0a0 | jens-daniel-mueller | 2024-05-28 | Build site. |
| html | d533f68 | jens-daniel-mueller | 2024-05-28 | Build site. |
| html | 555c1c8 | jens-daniel-mueller | 2024-05-27 | Build site. |
| html | 7b6f27c | jens-daniel-mueller | 2024-05-27 | Build site. |
| html | 4be90dd | jens-daniel-mueller | 2024-05-27 | Build site. |
| html | e1e0ccb | jens-daniel-mueller | 2024-05-27 | Build site. |
| html | 7013182 | jens-daniel-mueller | 2024-05-27 | Build site. |
| html | 97eff6a | jens-daniel-mueller | 2024-05-25 | Build site. |
| html | fe97ed3 | jens-daniel-mueller | 2024-05-25 | Build site. |
| html | ac8cc7e | jens-daniel-mueller | 2024-05-24 | Build site. |
| html | 12f6571 | jens-daniel-mueller | 2024-05-22 | Build site. |
| html | 7868a54 | jens-daniel-mueller | 2024-05-22 | Build site. |
| html | 571e2f8 | jens-daniel-mueller | 2024-05-22 | Build site. |
| html | 563345f | jens-daniel-mueller | 2024-05-21 | Build site. |
| html | 29e0ec4 | jens-daniel-mueller | 2024-05-21 | Build site. |
| html | 7c08e1c | jens-daniel-mueller | 2024-05-21 | Build site. |
| html | 1eefab2 | jens-daniel-mueller | 2024-05-21 | Build site. |
| html | 5af03d1 | jens-daniel-mueller | 2024-05-17 | Build site. |
| html | a29d870 | jens-daniel-mueller | 2024-05-16 | Build site. |
| html | dbc1fc6 | jens-daniel-mueller | 2024-05-16 | Build site. |
| html | aea0b99 | jens-daniel-mueller | 2024-05-16 | Build site. |
| html | 3310cf6 | jens-daniel-mueller | 2024-05-16 | Build site. |
| html | fcd728c | jens-daniel-mueller | 2024-05-16 | Build site. |
| html | 960912c | jens-daniel-mueller | 2024-05-16 | Build site. |
| html | b7d0689 | jens-daniel-mueller | 2024-05-15 | Build site. |
| html | 00ad9d5 | jens-daniel-mueller | 2024-05-15 | Build site. |
| html | 47f8868 | jens-daniel-mueller | 2024-05-15 | Build site. |
| html | 589243f | jens-daniel-mueller | 2024-05-15 | Build site. |
| html | 1e4c153 | jens-daniel-mueller | 2024-05-14 | Build site. |
| html | 009791f | jens-daniel-mueller | 2024-05-14 | Build site. |
| html | 8c96de4 | jens-daniel-mueller | 2024-05-08 | Build site. |
| html | 3fea035 | jens-daniel-mueller | 2024-05-08 | Build site. |
| html | 4b81eaf | jens-daniel-mueller | 2024-05-07 | Build site. |
| html | 60abdac | jens-daniel-mueller | 2024-04-23 | Build site. |
| html | e44a62b | jens-daniel-mueller | 2024-04-23 | Build site. |
| html | 7f9c687 | jens-daniel-mueller | 2024-04-23 | Build site. |
| html | 1ff6eb0 | jens-daniel-mueller | 2024-04-22 | Build site. |
| html | 9ecd92e | jens-daniel-mueller | 2024-04-22 | Build site. |
| html | 231f7cd | jens-daniel-mueller | 2024-04-17 | Build site. |
| html | f6e9707 | jens-daniel-mueller | 2024-04-17 | Build site. |
| html | ce4e2a6 | jens-daniel-mueller | 2024-04-17 | Build site. |
| html | a5911f0 | jens-daniel-mueller | 2024-04-17 | Build site. |
| html | 6709afa | jens-daniel-mueller | 2024-04-12 | Build site. |
| html | 58e3680 | jens-daniel-mueller | 2024-04-11 | Build site. |
| html | 238d229 | jens-daniel-mueller | 2024-04-11 | Build site. |
| html | dfcf790 | jens-daniel-mueller | 2024-04-11 | Build site. |
| html | 139bc97 | jens-daniel-mueller | 2024-04-11 | manual deletion of files |
| html | 2c6f421 | jens-daniel-mueller | 2024-04-11 | Build site. |
| html | 37ccea4 | jens-daniel-mueller | 2024-04-08 | Build site. |
| html | 19f40c9 | jens-daniel-mueller | 2024-04-05 | Build site. |
| Rmd | 2336de2 | jens-daniel-mueller | 2024-04-05 | ensemble mean estimates added |
| html | 2793f67 | jens-daniel-mueller | 2024-04-05 | Build site. |
| html | 69dc18c | jens-daniel-mueller | 2024-04-04 | Build site. |
| html | c9d994c | jens-daniel-mueller | 2024-04-04 | Build site. |
| html | 40cb158 | jens-daniel-mueller | 2024-04-03 | Build site. |
| html | 7bb6113 | jens-daniel-mueller | 2024-04-03 | Build site. |
| Rmd | 7be0147 | jens-daniel-mueller | 2024-04-03 | trend maps included |
| html | 27b48a1 | jens-daniel-mueller | 2024-04-03 | Build site. |
| html | a83c8fc | jens-daniel-mueller | 2024-04-03 | Build site. |
| Rmd | 53c89bd | jens-daniel-mueller | 2024-04-03 | rebuild entire website incl 2022 anomalies |
center <- -160
boundary <- center + 180
target_crs <- paste0("+proj=robin +over +lon_0=", center)
# target_crs <- paste0("+proj=eqearth +over +lon_0=", center)
# target_crs <- paste0("+proj=eqearth +lon_0=", center)
# target_crs <- paste0("+proj=igh_o +lon_0=", center)
worldmap <- ne_countries(scale = 'small',
type = 'map_units',
returnclass = 'sf')
worldmap <- worldmap %>% st_break_antimeridian(lon_0 = center)
worldmap_trans <- st_transform(worldmap, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans)
coastline <- ne_coastline(scale = 'small', returnclass = "sf")
coastline <- st_break_antimeridian(coastline, lon_0 = 200)
coastline_trans <- st_transform(coastline, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col="grey") +
# geom_sf(data = coastline_trans)
bbox <- st_bbox(c(xmin = -180, xmax = 180, ymax = 65, ymin = -78), crs = st_crs(4326))
bbox <- st_as_sfc(bbox)
bbox_trans <- st_break_antimeridian(bbox, lon_0 = center)
bbox_graticules <- st_graticule(
x = bbox_trans,
crs = st_crs(bbox_trans),
datum = st_crs(bbox_trans),
lon = c(20, 20.001),
lat = c(-78,65),
ndiscr = 1e3,
margin = 0.001
)
bbox_graticules_trans <- st_transform(bbox_graticules, crs = target_crs)
rm(worldmap, coastline, bbox, bbox_trans)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col="grey") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans)
lat_lim <- ext(bbox_graticules_trans)[c(3,4)]*1.002
lon_lim <- ext(bbox_graticules_trans)[c(1,2)]*1.005
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey90", col = "grey90") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans, linewidth = 1) +
# coord_sf(crs = target_crs,
# ylim = lat_lim,
# xlim = lon_lim,
# expand = FALSE) +
# theme(
# panel.border = element_blank(),
# axis.text = element_blank(),
# axis.ticks = element_blank()
# )
latitude_graticules <- st_graticule(
x = bbox_graticules,
crs = st_crs(bbox_graticules),
datum = st_crs(bbox_graticules),
lon = c(20, 20.001),
lat = c(-60,-30,0,30,60),
ndiscr = 1e3,
margin = 0.001
)
latitude_graticules_trans <- st_transform(latitude_graticules, crs = target_crs)
latitude_labels <- data.frame(lat_label = c("60°N","30°N","Eq.","30°S","60°S"),
lat = c(60,30,0,-30,-60)-4, lon = c(35)-c(0,2,4,2,0))
latitude_labels <- st_as_sf(x = latitude_labels,
coords = c("lon", "lat"),
crs = "+proj=longlat")
latitude_labels_trans <- st_transform(latitude_labels, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col = "grey") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans) +
# geom_sf(data = latitude_graticules_trans,
# col = "grey60",
# linewidth = 0.2) +
# geom_sf_text(data = latitude_labels_trans,
# aes(label = lat_label),
# size = 3,
# col = "grey60")pco2_product_list <- c("OceanSODAv2", "SOM-FFN", "fCO2-Residual", "CMEMS")
gobm_product_list <- c("ETHZ-CESM", "FESOM-REcoM")
files <- list.files(here::here("data/"), pattern = "FESOM-REcoM")
file_types <- str_remove(files, "FESOM-REcoM_2023_")
GCB_products = FALSEpCO2_product_synopsis <-
knitr::knit_expand(
file = here::here("analysis/child/pCO2_product_synopsis.Rmd"),
year_anom = 2023
)Read data
map <-
read_rds(here::here("data/map.rds"))
key_biomes <-
read_rds(here::here("data/key_biomes.rds"))
key_biomes <-
key_biomes[!str_detect(key_biomes, "NP")]
biome_mask <-
read_rds(here::here("data/biome_mask.rds"))
biome_mask_print <-
biome_mask %>%
filter(!str_detect(biome, "SO-SPSS|SO-ICE|Arctic")) %>%
# filter(!str_detect(biome, "SO-ICE|Arctic")) %>%
select(lon, lat)
region_biomes <-
read_rds(here::here("data/region_biomes.rds"))nino_sst <- read_table(here::here("data/nino34sst.txt"))
nino_sst <-
nino_sst %>%
select(year = YR,
month = MON,
resid = ANOM_3)co2_annmean_gl <- read_csv(here::here("data/co2_annmean_gl.csv"),
skip = 37)
co2_gr_gl <- read_csv(here::here("data/co2_gr_gl.csv"),
skip = 43)name_core <- c("fgco2", "fgco2_int", "fgco2_hov",
# "sfco2", "atm_fco2",
"dfco2",
# "kw_sol",
"temperature",
# "salinity",
# "dissic", "talk", "sdissic", "stalk", "cstar",
"sdissic_stalk",
"no3", "o2",
"mld", "thetao",
# "so",
"intpp", "chl",
"sfco2_therm","sfco2_nontherm","sfco2_total",
"resid_fgco2_dfco2", "resid_fgco2_kw_sol", "resid_fgco2_dfco2_kw_sol")
all_product_list <- c(pco2_product_list, gobm_product_list)
color_products <- c(
"OceanSODAv2" = "#672933",
"OceanSODA" = "#672933",
"SOM-FFN" = "#d1495b",
"fco2residual" = "#edae49",
"fCO2-Residual" = "#edae49",
"LDEO-HPD" = "#edae49",
"CMEMS" = "#AD8E55",
"ETHZ-CESM" = "#66a182",
"FESOM-REcoM" = "#00798c",
"CSIR-ML6" = "grey10",
"JMA-MLR" = "grey30",
"NIES-ML3" = "grey50",
"UExP-FNN-U" = "grey70"
)
shape_products <- c(
"OceanSODAv2" = 0,
"OceanSODA" = 0,
"SOM-FFN" = 1,
"fco2residual" = 2,
"fCO2-Residual" = 2,
"LDEO-HPD" = 2,
"CMEMS" = 5,
"ETHZ-CESM" = 0,
"FESOM-REcoM" = 1,
"CSIR-ML6" = 6,
"JMA-MLR" = 7,
"NIES-ML3" = 9,
"UExP-FNN-U" = 10
)
warm_color <- "#c33c57"
cold_color <- "#3f6fb3"
trend_color <- "#66a182"
warm_cool_gradient <-
rev(c(
"#61195a",
"#6f185f",
"#8d1e62",
"#aa2960",
"#c33c57",
"#da5351",
"#e77155",
"#f09264",
"#f09264",
"#fbd297",
"#fefefe",
"#c6e8ea",
"#97d4db",
"#79bcd0",
"#5ca2c6",
"#4a88bc",
"#3f6fb3",
"#3e56a2",
"#3c3f82",
"#2f2c5a",
"#272648"
))
# cmocean("balance")(100)for(i_file_type in file_types) {
# print(i_file_type)
# i_file_type <- file_types[1]
files <- list.files(here::here("data"),
pattern = paste(2023, i_file_type, sep = "_"),
full.names = TRUE)
if (GCB_products) {
files <- str_subset(files, paste("_GCB_", paste(gobm_product_list, collapse = "|"), sep = "|"))
files <- str_subset(files, paste(
paste(pco2_product_list, collapse = "|"),
paste(gobm_product_list, collapse = "|"),
sep = "|"
))
} else {
files <- str_subset(files, "_GCB_", negate = TRUE)
}
pco2_product <-
read_csv(files, id = "product")
# pco2_product %>%
# distinct(product)
pco2_product <-
pco2_product %>%
mutate(
product = str_extract(
product,
paste(all_product_list, collapse = "|")
)
)
if (!str_detect(files[1], "slope|_temperature_predict")) {
pco2_product <-
pco2_product %>%
mutate(
name = factor(name, levels = name_core),
product = factor(product, levels = all_product_list)
) %>%
filter(!is.na(name))
} else {
pco2_product <-
pco2_product %>%
mutate(product = factor(product, levels = all_product_list))
}
i_file_type <- str_remove(i_file_type, ".csv")
assign(paste("pco2_product", i_file_type, sep = "_"), pco2_product)
}Define labels and breaks
labels_breaks <- function(i_name) {
if (i_name == "dco2") {
i_legend_title <- "ΔpCO<sub>2</sub> anom.<br>(µatm)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "dfco2") {
i_legend_title <- "ΔfCO<sub>2</sub> anom.<br>(µatm)"
i_breaks <- c(-Inf, seq(-12, 12, 3), Inf)
}
if (i_name == "atm_co2") {
i_legend_title <- "pCO<sub>2,atm</sub> anom.<br>(µatm)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "atm_fco2") {
i_legend_title <- "fCO<sub>2,atm</sub> anom.<br>(µatm)"
i_breaks <- c(-Inf, seq(-2, 2, 0.5), Inf)
}
if (i_name == "sol") {
i_legend_title <- "K<sub>0</sub> anom.<br>(mol m<sup>-3</sup> µatm<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "kw") {
i_legend_title <- "k<sub>w</sub> anom.<br>(m yr<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "kw_sol") {
i_legend_title <- "k<sub>w</sub> K<sub>0</sub> anom.<br>(mol yr<sup>-1</sup> m<sup>-2</sup> µatm<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.015, 0.015, 0.003), Inf)
}
if (i_name == "spco2") {
i_legend_title <- "pCO<sub>2,ocean</sub> anom.<br>(µatm)"
i_breaks <- c(-Inf, seq(-12, 12, 3), Inf)
}
if (i_name == "sfco2") {
i_legend_title <- "fCO<sub>2,ocean</sub> anom.<br>(µatm)"
i_breaks <- c(-Inf, seq(-12, 12, 3), Inf)
}
if (i_name == "intpp") {
i_legend_title <- "NPP<sub>int</sub> anom.<br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-3, 3, 0.5), Inf)
}
if (i_name == "no3") {
i_legend_title <- "NO<sub>3</sub> anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-1.5, 1.5, 0.3), Inf)
}
if (i_name == "o2") {
i_legend_title <- "O<sub>2</sub> anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "dissic") {
i_legend_title <- "DIC anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-15, 15, 3), Inf)
}
if (i_name == "sdissic") {
i_legend_title <- "sDIC anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-15, 15, 3), Inf)
}
if (i_name == "cstar") {
i_legend_title <- "C* anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "talk") {
i_legend_title <- "TA anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-15, 15, 3), Inf)
}
if (i_name == "stalk") {
i_legend_title <- "sTA anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-15, 15, 3), Inf)
}
if (i_name == "sdissic_stalk") {
i_legend_title <- "sDIC - sTA anom.<br>(μmol kg<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-15, 15, 3), Inf)
}
if (i_name == "sfco2_total") {
i_legend_title <- "total"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "sfco2_therm") {
i_legend_title <- "thermal"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "sfco2_nontherm") {
i_legend_title <- "non-thermal"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "fgco2") {
i_legend_title <- "FCO<sub>2</sub> anom.<br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "slope") {
i_legend_title <- "Slope FCO<sub>2</sub> anom. / SST anom.<br>(mol m<sup>-2</sup> yr<sup>-1</sup> °C<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-1, 1, 0.25), Inf)
}
if (i_name == "fgco2_predict") {
i_legend_title <- "FCO<sub>2</sub> anom. pred.<br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "fgco2_hov") {
i_legend_title <- "FCO<sub>2</sub> anom.<br>(PgC deg<sup>-1</sup> yr<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "fgco2_int") {
i_legend_title <- "FCO<sub>2</sub> anom.<br>(PgC yr<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "fgco2_predict_int") {
i_legend_title <- "FCO<sub>2</sub> anom. pred.<br>(PgC yr<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "thetao") {
i_legend_title <- "Temp. anom.<br>(°C)"
i_breaks <- c(-Inf, seq(-1.6, 1.6, 0.4), Inf)
}
if (i_name == "temperature") {
i_legend_title <- "SST anom.<br>(°C)"
i_breaks <- c(-Inf, seq(-1.6, 1.6, 0.4), Inf)
}
if (i_name == "salinity") {
i_legend_title <- "SSS anom."
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "so") {
i_legend_title <- "Salinity anom."
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "chl") {
i_legend_title <- "lg(Chl-a) anom.<br>(lg(mg m<sup>-3</sup>))"
i_breaks <- c(-Inf, seq(-0.2, 0.2, 0.05), Inf)
}
if (i_name == "mld") {
i_legend_title <- "MLD anom.<br>(m)"
i_breaks <- c(-Inf, seq(-40, 40, 10), Inf)
}
if (i_name == "press") {
i_legend_title <- "pressure<sub>atm</sub> anom.<br>(Pa)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "wind") {
i_legend_title <- "Wind anom.<br>(m sec<sup>-1</sup>)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "SSH") {
i_legend_title <- "SSH anom.<br>(m)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "fice") {
i_legend_title <- "Sea ice anom.<br>(%)"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "resid_fgco2") {
i_legend_title <-
"Observed"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "resid_fgco2_dfco2") {
i_legend_title <-
"ΔfCO<sub>2</sub> contr."
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "resid_fgco2_kw_sol") {
i_legend_title <-
"k<sub>w</sub> K<sub>0</sub> contr."
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "resid_fgco2_dfco2_kw_sol") {
i_legend_title <-
"ΔfCO<sub>2</sub> ⨯ k<sub>w</sub> K<sub>0</sub> contr."
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "resid_fgco2_sum") {
i_legend_title <-
"∑"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
if (i_name == "resid_fgco2_offset") {
i_legend_title <-
"Obs. - ∑"
i_breaks <- c(-Inf, seq(-0.5, 0.5, 0.1), Inf)
}
all_labels_breaks <- lst(i_legend_title, i_breaks)
return(all_labels_breaks)
}
x_axis_labels <-
c(
"dco2" = labels_breaks("dco2")$i_legend_title,
"dfco2" = labels_breaks("dfco2")$i_legend_title,
"atm_co2" = labels_breaks("atm_co2")$i_legend_title,
"atm_fco2" = labels_breaks("atm_fco2")$i_legend_title,
"sol" = labels_breaks("sol")$i_legend_title,
"kw" = labels_breaks("kw")$i_legend_title,
"kw_sol" = labels_breaks("kw_sol")$i_legend_title,
"intpp" = labels_breaks("intpp")$i_legend_title,
"no3" = labels_breaks("no3")$i_legend_title,
"o2" = labels_breaks("o2")$i_legend_title,
"dissic" = labels_breaks("dissic")$i_legend_title,
"sdissic" = labels_breaks("sdissic")$i_legend_title,
"cstar" = labels_breaks("cstar")$i_legend_title,
"talk" = labels_breaks("talk")$i_legend_title,
"stalk" = labels_breaks("stalk")$i_legend_title,
"sdissic_stalk" = labels_breaks("sdissic_stalk")$i_legend_title,
"spco2" = labels_breaks("spco2")$i_legend_title,
"sfco2" = labels_breaks("sfco2")$i_legend_title,
"sfco2_total" = labels_breaks("sfco2_total")$i_legend_title,
"sfco2_therm" = labels_breaks("sfco2_therm")$i_legend_title,
"sfco2_nontherm" = labels_breaks("sfco2_nontherm")$i_legend_title,
"fgco2" = labels_breaks("fgco2")$i_legend_title,
"slope" = labels_breaks("slope")$i_legend_title,
"fgco2_predict" = labels_breaks("fgco2_predict")$i_legend_title,
"fgco2_hov" = labels_breaks("fgco2_hov")$i_legend_title,
"fgco2_int" = labels_breaks("fgco2_int")$i_legend_title,
"fgco2_predict_int" = labels_breaks("fgco2_predict_int")$i_legend_title,
"thetao" = labels_breaks("thetao")$i_legend_title,
"temperature" = labels_breaks("temperature")$i_legend_title,
"salinity" = labels_breaks("salinity")$i_legend_title,
"so" = labels_breaks("so")$i_legend_title,
"chl" = labels_breaks("chl")$i_legend_title,
"mld" = labels_breaks("mld")$i_legend_title,
"press" = labels_breaks("press")$i_legend_title,
"wind" = labels_breaks("wind")$i_legend_title,
"SSH" = labels_breaks("SSH")$i_legend_title,
"fice" = labels_breaks("fice")$i_legend_title,
"resid_fgco2" = labels_breaks("resid_fgco2")$i_legend_title,
"resid_fgco2_dfco2" = labels_breaks("resid_fgco2_dfco2")$i_legend_title,
"resid_fgco2_kw_sol" = labels_breaks("resid_fgco2_kw_sol")$i_legend_title,
"resid_fgco2_dfco2_kw_sol" = labels_breaks("resid_fgco2_dfco2_kw_sol")$i_legend_title,
"resid_fgco2_sum" = labels_breaks("resid_fgco2_sum")$i_legend_title,
"resid_fgco2_offset" = labels_breaks("resid_fgco2_offset")$i_legend_title
)
# create axis labels for absolute values by removing anom.
x_axis_labels_abs <- x_axis_labels
x_axis_labels_abs <- str_replace_all(x_axis_labels_abs, " anom.", "")
names(x_axis_labels_abs) <- names(x_axis_labels)Functions
Seasonality plots
p_season <- function(df,
dim_row = "name",
dim_col = "product",
title = NULL,
var = "resid",
scales = "free_y") {
p <- ggplot(data = df,
aes(month, !!ensym(var)))
if(var == "resid"){
p <- p +
geom_hline(yintercept = 0, linewidth =0.5)
}
p <- p +
geom_path(data = . %>% filter(year != 2023),
aes(group = as.factor(year),
col = as.factor(paste(min(year), max(year), sep = "-"))),
alpha = 0.5)+
geom_path(data = . %>%
filter(year != 2023) %>%
group_by_at(vars(month, dim_col, dim_row)) %>%
summarise(!!ensym(var) := mean(!!ensym(var))),
aes(col = "Climatological\nmean"),
linewidth = 0.7) +
scale_color_manual(values = c("grey60", "grey10"),
guide = guide_legend(order = 2,
reverse = TRUE)) +
new_scale_color()+
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(year)),
linewidth = 1.2) +
scale_color_manual(
values = warm_color,
guide = guide_legend(order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = title,
x = "Month")
if(df %>% filter(name == "fgco2") %>% nrow() > 0 & "value" %in% names(df)){
df_sink <- df %>%
filter(year == 2023,
name == "fgco2")
p <- p +
geom_point(data = df_sink %>% filter(value < 0),
aes(shape = "Sink"), fill = "white") +
geom_point(data = df_sink %>% filter(value >= 0),
aes(shape = "Source"), fill = "white") +
scale_shape_manual(values = c(25,24))
}
if (!(is.null(dim_col))) {
p <- p +
facet_grid2(
as.formula(paste(dim_row, "~", dim_col)),
scales = scales,
# independent = "y",
labeller = labeller(name = x_axis_labels),
switch = "y"
)
} else {
p <- p +
facet_grid(
as.formula(paste(dim_row, "~ .")),
scales = scales,
# independent = "y",
labeller = labeller(name = x_axis_labels),
switch = "y"
)
}
p <- p +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank(),
axis.text.y.right = element_blank()
)
# scale_y_continuous(sec.axis = dup_axis())
p
}fCO2 decomposition
fco2_decomposition <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
# group_by <- quos(biome, year, month)
pco2_product_biome_monthly_fCO2_decomposition <-
df %>%
filter(name %in% c("temperature", "sfco2"))
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "temperature") %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid),
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "sfco2") %>%
select(-c(value, resid)) %>%
pivot_wider(values_from = fit)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(sfco2_therm = (sfco2 * exp(0.0423 * temperature)) - sfco2)
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition,
df %>%
filter(name %in% c("sfco2")) %>%
select(-c(value, fit, name)) %>%
rename(sfco2_total = resid)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(sfco2_nontherm = sfco2_total - sfco2_therm)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
select(-c(temperature, sfco2)) %>%
pivot_longer(starts_with("sfco2"),
values_to = "resid")
}Flux attribution
flux_attribution <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
pco2_product_flux_attribution <-
df %>%
filter(name %in% c("dfco2", "kw_sol", "fgco2"))
pco2_product_flux_attribution <-
inner_join(
pco2_product_flux_attribution %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid,
names_prefix = "resid_"),
pco2_product_flux_attribution %>%
select(-c(value, resid)) %>%
filter(name != "fgco2") %>%
pivot_wider(values_from = fit)
)
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
mutate(
resid_fgco2_dfco2 = resid_dfco2 * kw_sol,
resid_fgco2_kw_sol = resid_kw_sol * dfco2,
resid_fgco2_dfco2_kw_sol = resid_dfco2 * resid_kw_sol
# resid_fgco2_sum = resid_fgco2_dfco2 + resid_fgco2_kw_sol + resid_fgco2_dfco2_kw_sol
)
# pco2_product_flux_attribution <-
# pco2_product_flux_attribution %>%
# mutate(resid_fgco2_offset = resid_fgco2 - resid_fgco2_sum)
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
select(product, !!!group_by, starts_with("resid_fgco2")) %>%
pivot_longer(starts_with("resid_"),
values_to = "resid")
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
filter(str_detect(name, "dfco2|kw_sol")) %>%
mutate(name = factor(
name,
levels = c(
"resid_fgco2",
"resid_fgco2_dfco2",
"resid_fgco2_kw_sol",
"resid_fgco2_dfco2_kw_sol",
"resid_fgco2_sum",
"resid_fgco2_offset"
)
))
}Robinson map
bbox <- st_bbox(c(xmin = -180, xmax = 180, ymax = 76, ymin = -54), crs = st_crs(4326))
# bbox <- st_bbox(c(xmin = -180, xmax = 180, ymax = 85, ymin = -80), crs = st_crs(4326))
bbox <- st_as_sfc(bbox)
bbox_trans <- st_break_antimeridian(bbox, lon_0 = center)
bbox_graticules <- st_graticule(
x = bbox_trans,
crs = st_crs(bbox_trans),
datum = st_crs(bbox_trans),
lon = c(20, 20.001),
lat = c(-54,76),
# lat = c(-80,85),
ndiscr = 1e3,
margin = 0.001
)
bbox_graticules_trans <- st_transform(bbox_graticules, crs = target_crs)
rm(bbox, bbox_trans)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col="grey") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans)
lat_lim <- ext(bbox_graticules_trans)[c(3,4)]*1.002
lon_lim <- ext(bbox_graticules_trans)[c(1,2)]*1.005
p_map_mdim_robinson <-
function(df,
df_uncertainty = NULL,
dim_row = NULL,
dim_col = NULL,
dim_wrap = NULL,
n_col = NULL,
var,
legend_title = NULL,
breaks = NULL,
n_labels = 2,
target_crs = "+proj=robin +over +lon_0=-160",
col = "divergent",
col_scale = "warm_cold",
plot_latitudes = FALSE,
legend_position = "top") {
if (is.null(dim_col) & is.null(dim_row) & is.null(dim_wrap)) {
df_raster <- df %>%
select(lon, lat, all_of(var)) %>%
rast(crs = "+proj=longlat")
df_raster <-
project(df_raster, target_crs)
df_tibble <-
df_raster %>%
as.data.frame(xy = TRUE, na.rm = FALSE) %>%
as_tibble() %>%
rename(lon = x, lat = y) %>%
drop_na()
} else {
# if (!is.null(dim_col) & !is.null(dim_row) & !is.null(dim_wrap)) {
# names_sep <- ";"
# } else {
# names_sep <- NULL
# }
names_sep <- ";"
df_raster <- df %>%
select(lon, lat,
all_of(c(dim_row, dim_col, dim_wrap)),
all_of(var)) %>%
pivot_wider(names_from = all_of(c(dim_row, dim_col, dim_wrap)),
values_from = all_of(var),
names_sep = names_sep) %>%
rast(crs = "+proj=longlat")
df_raster <-
project(df_raster, target_crs)
if (length(c(dim_row, dim_col, dim_wrap)) <= 1) {
names_sep <- NULL
}
df_tibble <-
df_raster %>%
as.data.frame(xy = TRUE, na.rm = FALSE) %>%
as_tibble() %>%
rename(lon = x, lat = y) %>%
pivot_longer(
-c(lon, lat),
names_sep = names_sep,
names_to = c(dim_row, dim_col, dim_wrap),
values_to = var
) %>%
drop_na()
}
if (is.null(legend_title)) {
legend_title <- var
}
var <- sym(var)
p_map <- ggplot() +
geom_raster(data = df_tibble, aes(
x = lon,
y = lat,
fill = cut(!!var, breaks, include.lowest = TRUE)
))
p_map <- p_map +
geom_sf(data = worldmap_trans %>% select(-name),
fill = "grey90",
col = "grey90") +
geom_sf(data = coastline_trans, linewidth = 0.3) +
geom_sf(data = bbox_graticules_trans, linewidth = 0.5)
if (plot_latitudes) {
p_map <- p_map +
geom_sf(data = latitude_graticules_trans,
col = "grey60",
linewidth = 0.2) +
geom_sf_text(
data = latitude_labels_trans,
aes(label = lat_label),
size = 3,
col = "grey60"
)
}
if (!is.null(df_uncertainty)) {
p_map <- p_map +
geom_sf(
data = df_uncertainty %>% filter(signif_single == 0),
col = "grey60",
size = 0.05
)
}
p_map <- p_map +
coord_sf(
crs = target_crs,
ylim = lat_lim,
xlim = lon_lim,
expand = FALSE
)
if (legend_position == "top") {
p_map <- p_map +
guides(
fill = guide_colorsteps(
barheight = unit(0.3, "cm"),
barwidth = unit(8, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme_void() +
theme(
legend.margin=margin(t = .1, b = .1, unit='cm'),
plot.margin = margin(.1,.1,.1,.1,"cm"),
panel.spacing = unit(.1,"cm"),
legend.position = "top",
legend.title.align = 1,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(halign = 1, lineheight = 1.5)
)
}
if (legend_position == "bottom") {
p_map <- p_map +
guides(
fill = guide_colorsteps(
barheight = unit(0.3, "cm"),
barwidth = unit(8, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "bottom",
direction = "horizontal"
)
) +
theme_void() +
theme(
legend.margin=margin(t = .1, b = .1, unit='cm'),
plot.margin = margin(.1,.1,.1,.1,"cm"),
panel.spacing = unit(.1,"cm"),
legend.position = "bottom",
legend.title.align = 1,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(halign = 1, lineheight = 1.5)
)
}
if (legend_position == "right") {
p_map <- p_map +
guides(
fill = guide_colorsteps(
barheight = unit(6, "cm"),
barwidth = unit(0.3, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "right",
direction = "vertical"
)
) +
theme_void() +
theme(
legend.position = "right",
legend.title.align = 0,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(halign = 0, lineheight = 1.5)
)
}
if (legend_position == "left") {
p_map <- p_map +
guides(
fill = guide_colorsteps(
barheight = unit(6, "cm"),
barwidth = unit(0.3, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "left",
direction = "vertical"
)
) +
theme_void() +
theme(
legend.position = "left",
legend.title.align = 0,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(halign = 0, lineheight = 1.5)
)
}
if (col == "sequential") {
breaks_test <- breaks[!breaks == Inf]
breaks_test <- breaks_test[!breaks_test == -Inf]
breaks_reverse <-
abs(first(breaks_test)) < abs(last(breaks_test))
if (breaks_reverse == TRUE) {
direction_value = 1
reverse_value = TRUE
} else{
direction_value = -1
reverse_value = FALSE
}
if (n_labels == 1) {
labels <- breaks_test
} else {
breaks_test[seq_along(breaks_test) %% 2 == 0] <- ""
labels <- breaks_test
}
if (col_scale %in% c("viridis", "plasma", "cividis")) {
p_map <- p_map +
scale_fill_viridis_d(
drop = FALSE,
name = legend_title,
direction = direction_value,
option = col_scale,
labels = unname(labels)
)
}
} else {
breaks_test <- breaks[!breaks == Inf]
breaks_test <- breaks_test[!breaks_test == -Inf]
if (n_labels == 1) {
labels <- breaks_test
} else {
breaks_test[seq_along(breaks_test) %% 2 == 0] <- ""
labels <- breaks_test
}
p_map <- p_map +
scale_fill_gradientn(
colours = warm_cool_gradient,
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
super = ScaleDiscretised,
name = legend_title,
labels = unname(labels)
)
# colorspace::scale_fill_discrete_divergingx(
# palette = "RdBu",
# drop = FALSE,
# rev = TRUE,
# name = legend_title,
# labels = unname(labels)
# )
}
if (!(is.null(dim_row) & is.null(dim_col))) {
if (is.null(dim_col)) {
dim_col <- "."
}
if (is.null(dim_row)) {
dim_row <- "."
}
p_map <- p_map +
facet_grid(as.formula(paste(dim_row, "~", dim_col)),
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(strip.text.x.top = element_markdown(),
strip.text.y.left = element_markdown())
}
if (!is.null(dim_wrap) & is.null(n_col)) {
p_map <- p_map +
facet_wrap(as.formula(paste("~", dim_wrap)))
}
if (!(is.null(dim_wrap) & is.null(n_col))) {
if (dim_wrap == "name") {
p_map <- p_map +
facet_wrap(as.formula(paste("~", dim_wrap)),
labeller = labeller(name = x_axis_labels),
ncol = n_col) +
theme(strip.text.x.top = element_markdown())
} else{
p_map <- p_map +
facet_wrap(as.formula(paste("~", dim_wrap)), ncol = n_col) +
theme(strip.text.x.top = element_markdown())
}
}
p_map
}Maps
The following maps show the anomalies of each variable in 2023 as provided through the fCO2 product. Anomalies are determined based on the predicted value of a linear regression model fit to the available data from 1990 to 2022.
Maps are first presented as annual means, and than as monthly means. Note that the 2023 predictions for the monthly maps are done individually for each month, such the mean seasonal anomaly from the annual mean is removed.
Note: The increase the computational speed, I regridded all maps to 5X5° grid.
Annual means
2023 anomaly
pco2_product_map_annual_anomaly <-
inner_join(
biome_mask_print,
pco2_product_map_annual_anomaly
)
pco2_product_map_annual_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(1) %>%
map(
~ p_map_mdim_robinson(
df = .x,
var = "resid",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
breaks = labels_breaks(.x %>% distinct(name))$i_breaks,
dim_wrap = "product",
n_col = 2
)
)[[1]]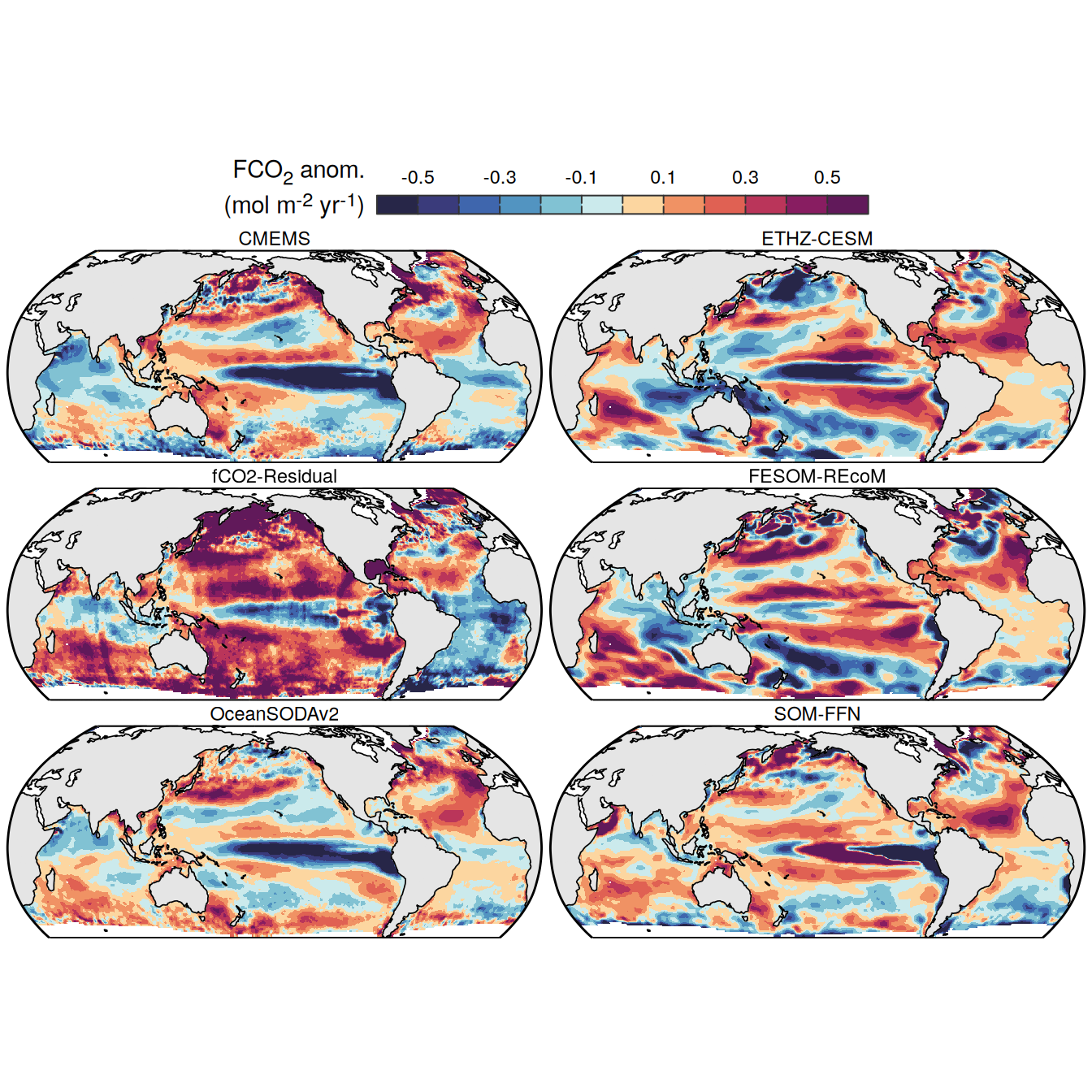
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[2]]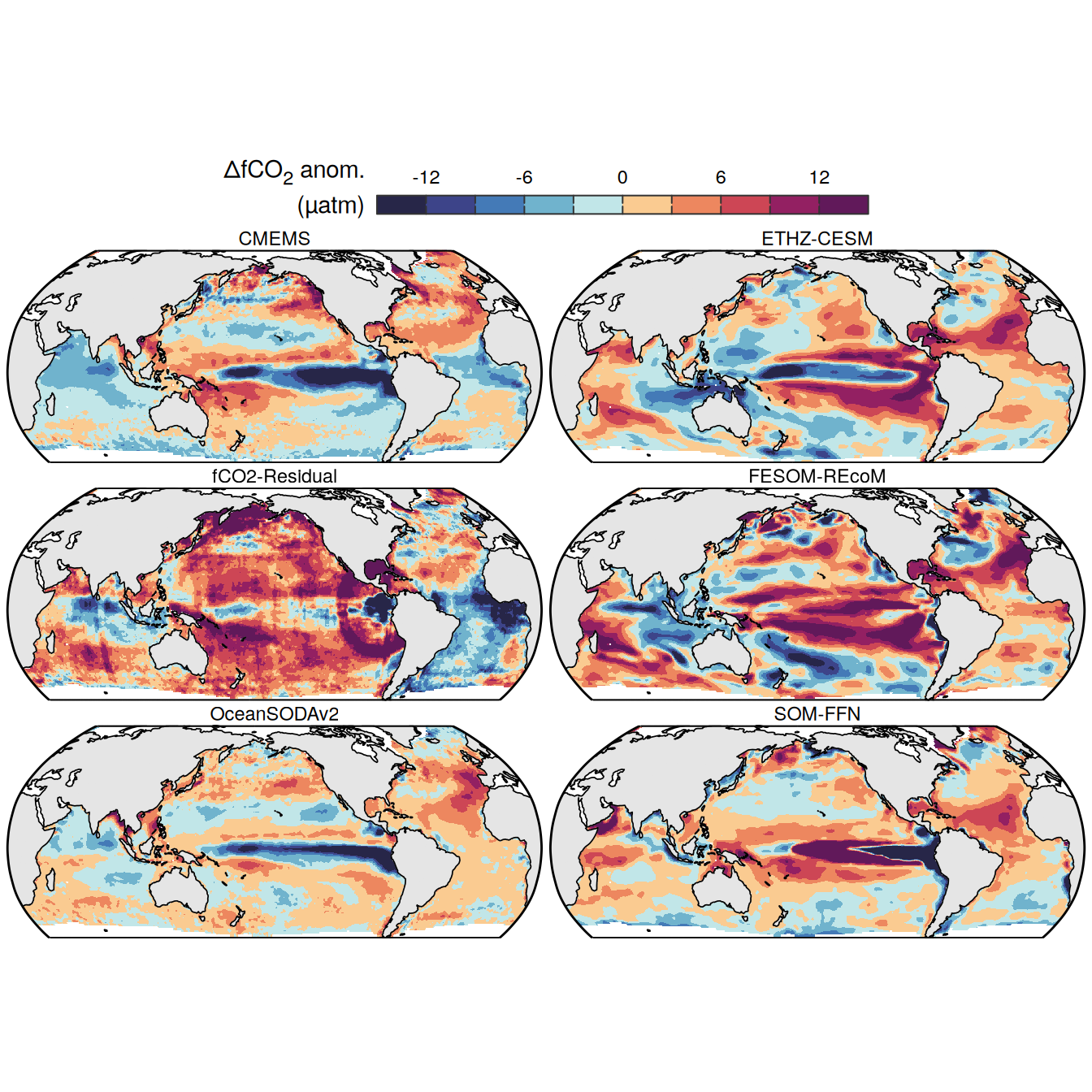
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[3]]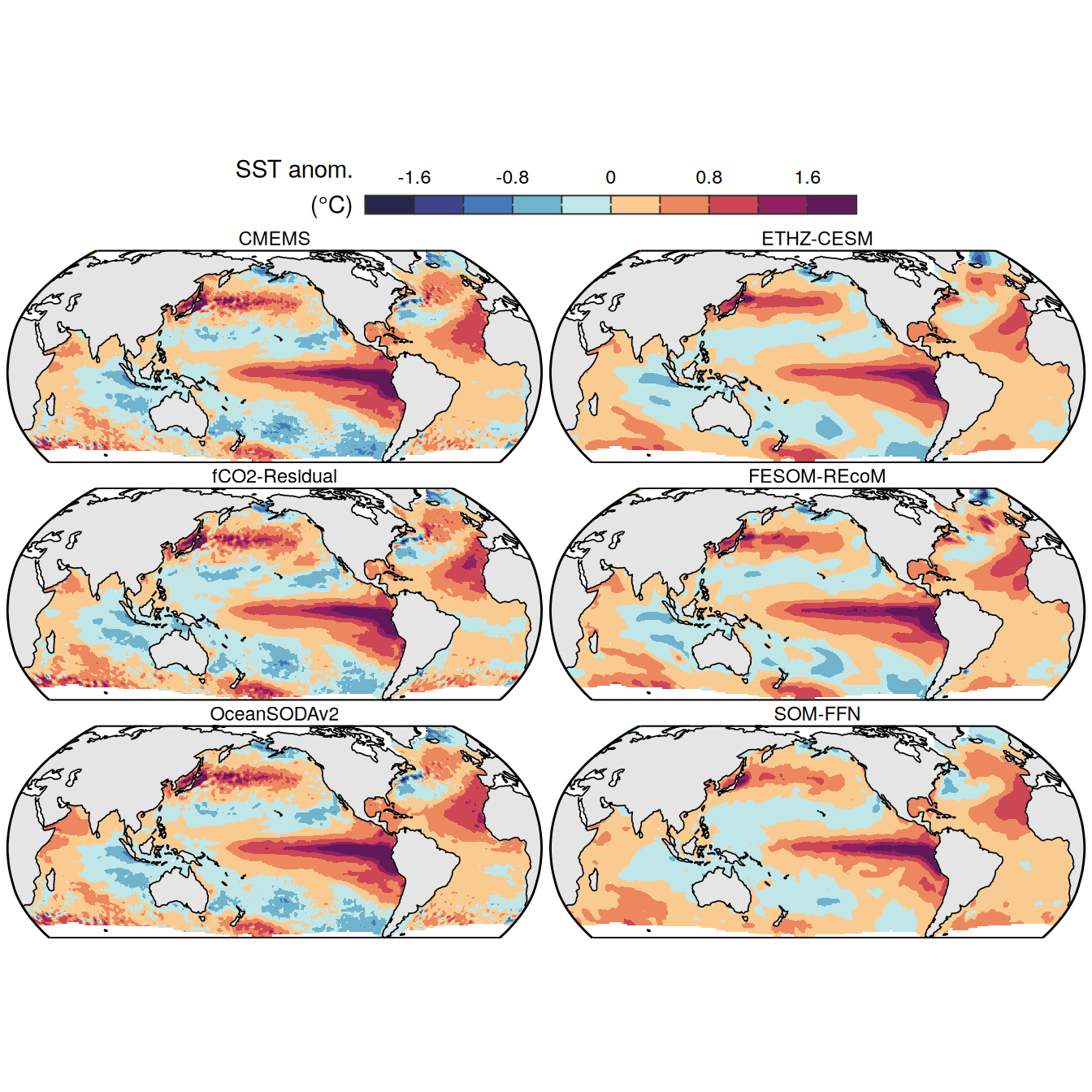
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[4]]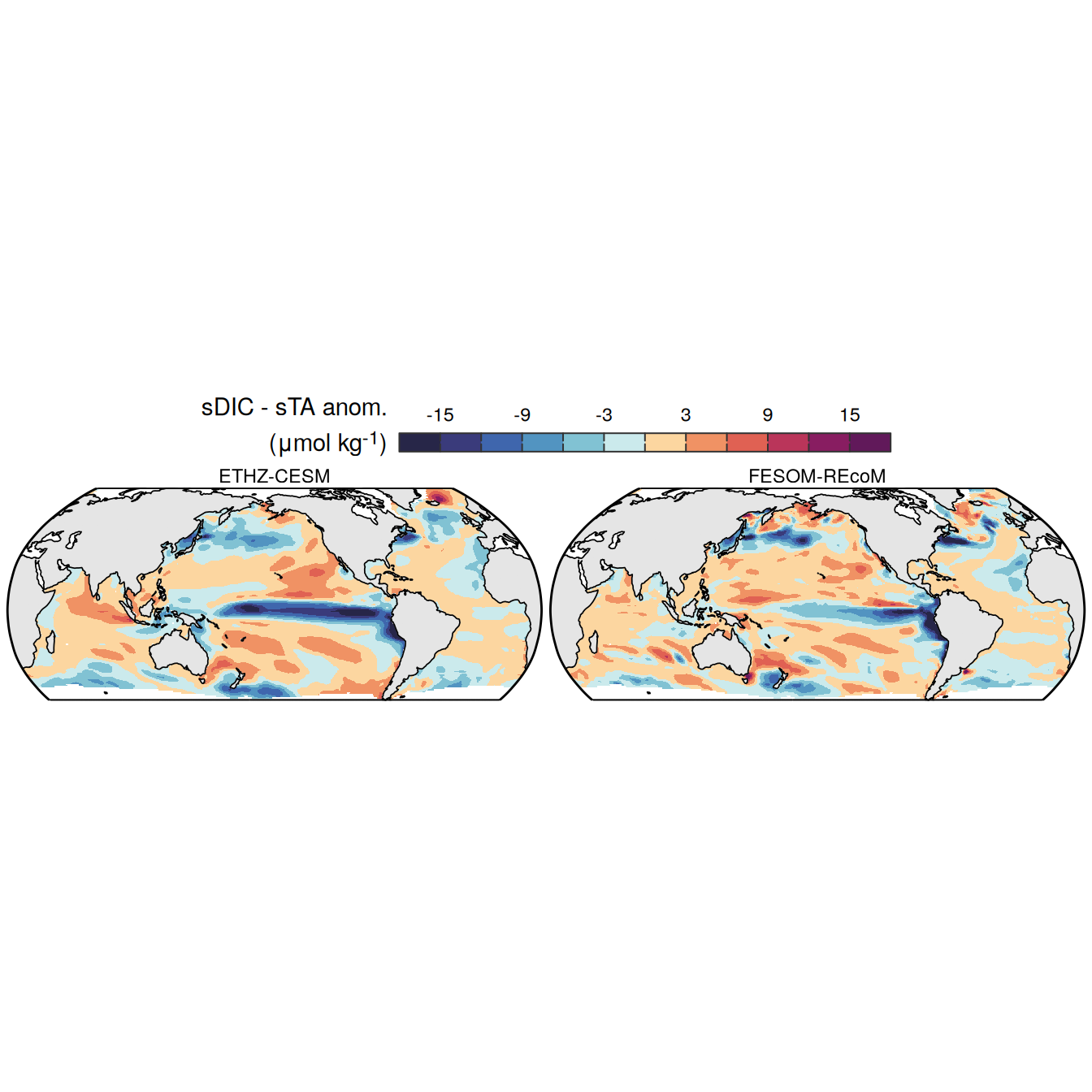
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[5]]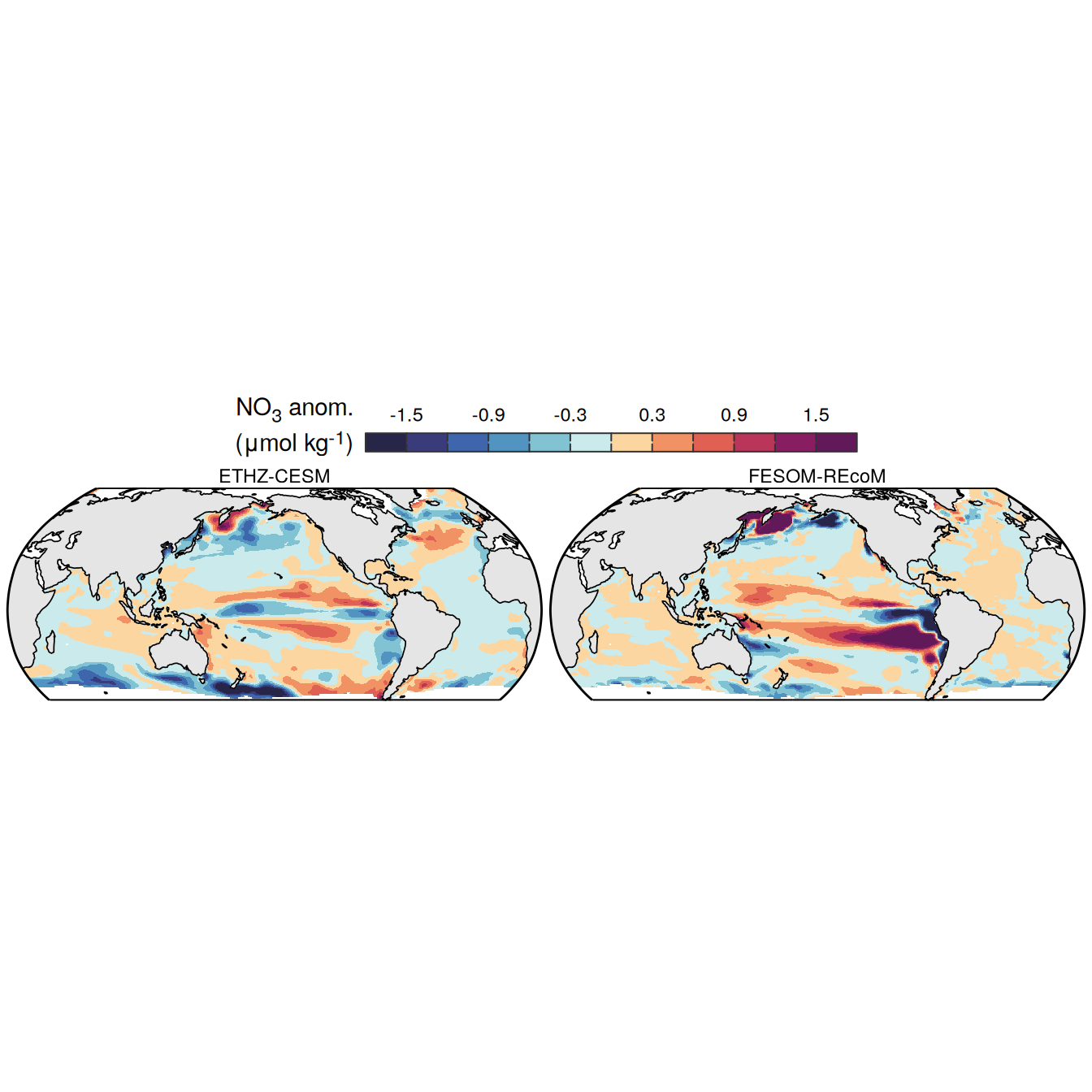
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[6]]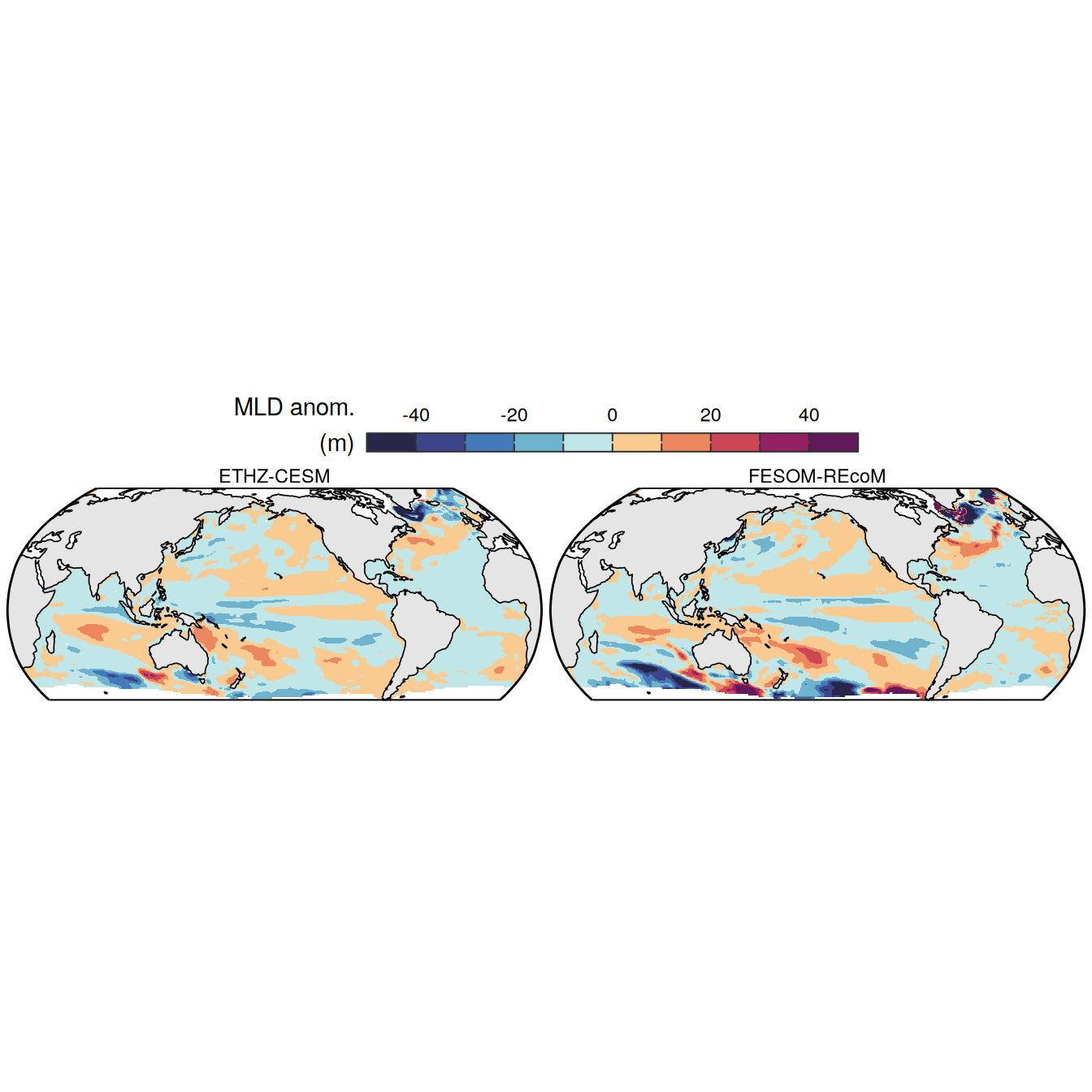
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[7]]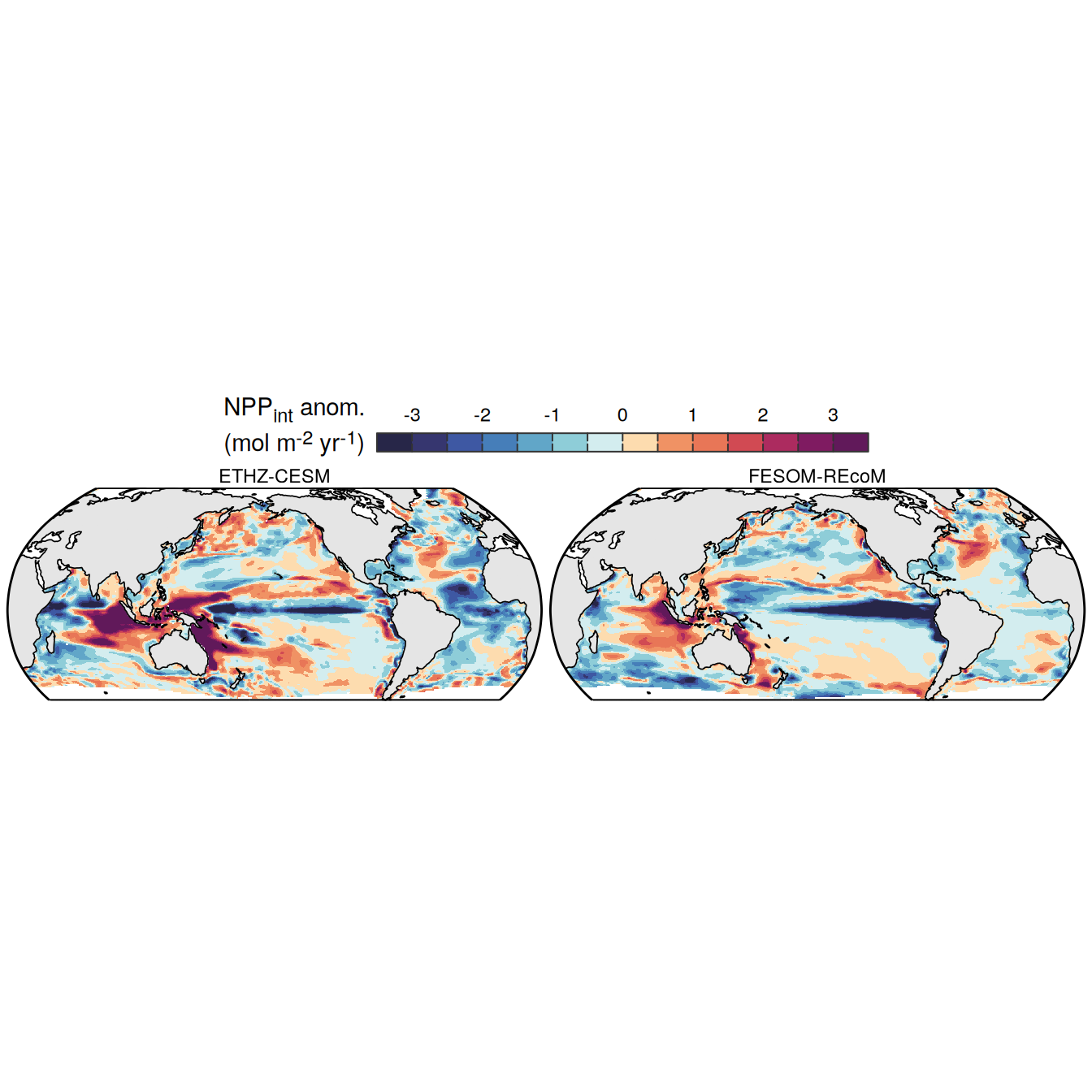
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[8]]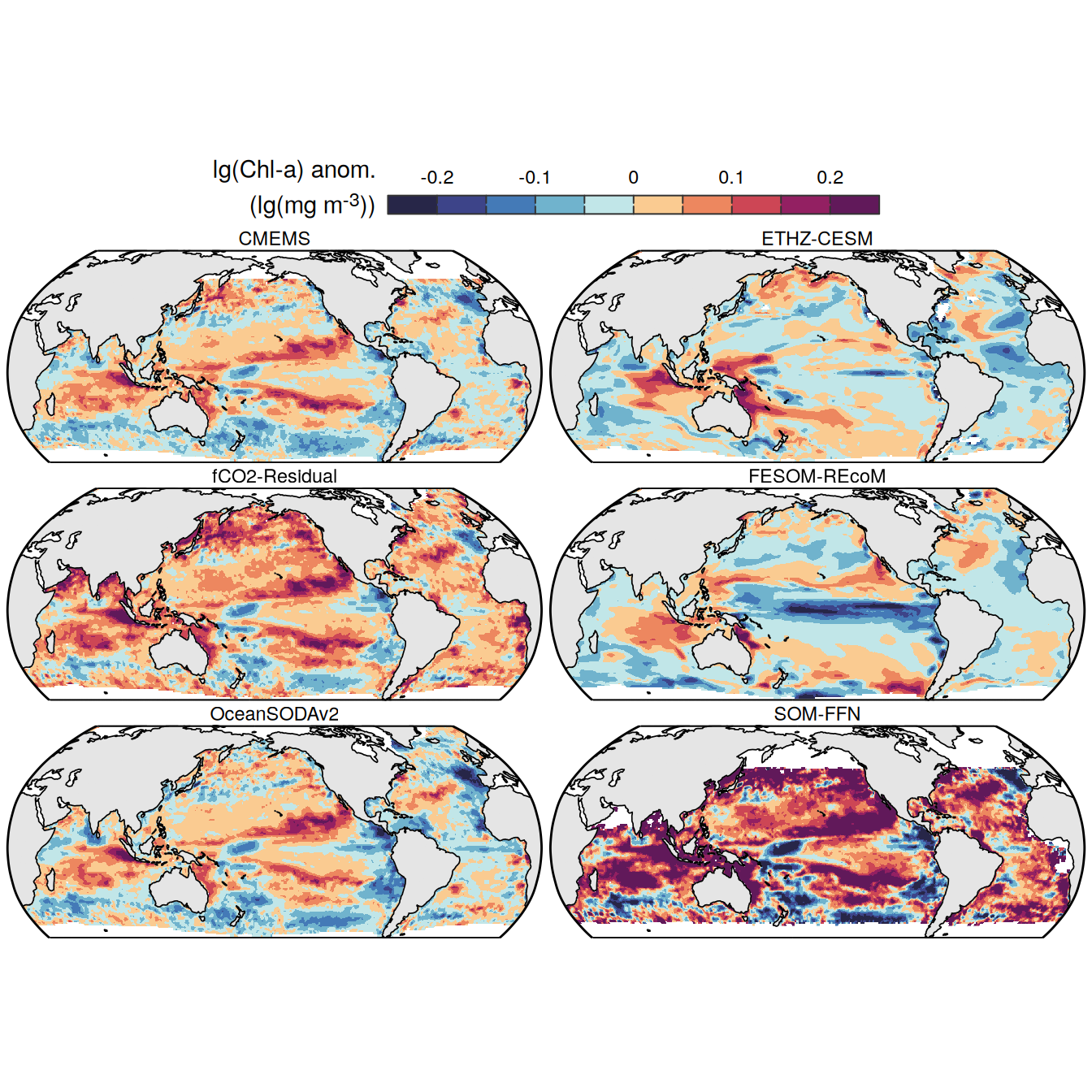
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
plot_list <-
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
product == "ETHZ-CESM",
name %in% c(
"fgco2",
"dfco2",
"kw_sol",
"temperature",
"salinity",
"sdissic",
"stalk",
"sdissic_stalk",
"no3",
"mld",
"intpp",
"chl"
)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ p_map_mdim_robinson(
df = .x,
var = "resid",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
breaks = labels_breaks(.x %>% distinct(name))$i_breaks
)
)
ggsave(plot = wrap_plots(plot_list,
ncol = 3,
byrow = FALSE),
width = 14,
height = 11,
filename = "../output/map_anomaly_ETHZ-CESM.jpg")plot_list <-
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
product == "FESOM-REcoM",
name %in% c(
"fgco2",
"dfco2",
"kw_sol",
"temperature",
"salinity",
"sdissic",
"stalk",
"sdissic_stalk",
"no3",
"mld",
"intpp",
"chl"
)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ p_map_mdim_robinson(
df = .x,
var = "resid",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
breaks = labels_breaks(.x %>% distinct(name))$i_breaks
)
)
ggsave(plot = wrap_plots(plot_list,
ncol = 3,
byrow = FALSE),
width = 14,
height = 11,
filename = "../output/map_anomaly_FESOM-REcoM.jpg")
rm(plot_list)pco2_product_map_annual_anomaly_ensemble <-
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
product %in% pco2_product_list) %>%
fgroup_by(name, lon, lat) %>%
fsummarise(
resid_sd = fsd(resid),
resid_mean = fmean(resid),
value_sd = fsd(value),
value_mean = fmean(value),
n = fnobs(resid)
) %>%
filter(n == length(pco2_product_list)) %>%
select(-n)
pco2_product_map_annual_anomaly_ensemble_coarse <-
m_grid_horizontal_coarse(pco2_product_map_annual_anomaly_ensemble) %>%
fgroup_by(name, lon_grid, lat_grid) %>%
fsummarise(
resid_sd_coarse = fmean(resid_sd, na.rm = TRUE),
resid_mean_coarse = fmean(resid_mean, na.rm = TRUE),
value_sd_coarse = fmean(value_sd, na.rm = TRUE),
value_mean_coarse = fmean(value_mean, na.rm = TRUE)
) %>%
rename(lon = lon_grid, lat = lat_grid)
pco2_product_map_annual_anomaly_ensemble_uncertainty <-
pco2_product_map_annual_anomaly_ensemble_coarse %>%
mutate(signif_single = if_else(abs(resid_mean_coarse) < resid_sd_coarse, 0, 1)) %>%
select(lon, lat, name, signif_single) %>%
st_as_sf(coords = c("lon", "lat"), crs = "+proj=longlat")
pco2_product_map_annual_anomaly_ensemble %>%
mutate(product = "Ensemble mean") %>%
group_split(name) %>%
# head(1) %>%
map(
~ p_map_mdim_robinson(
df = .x,
df_uncertainty = pco2_product_map_annual_anomaly_ensemble_uncertainty %>%
filter(name == .x %>% distinct(name) %>% pull()),
var = "resid_mean",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
breaks = labels_breaks(.x %>% distinct(name))$i_breaks,
n_labels = 2
)
)[[1]]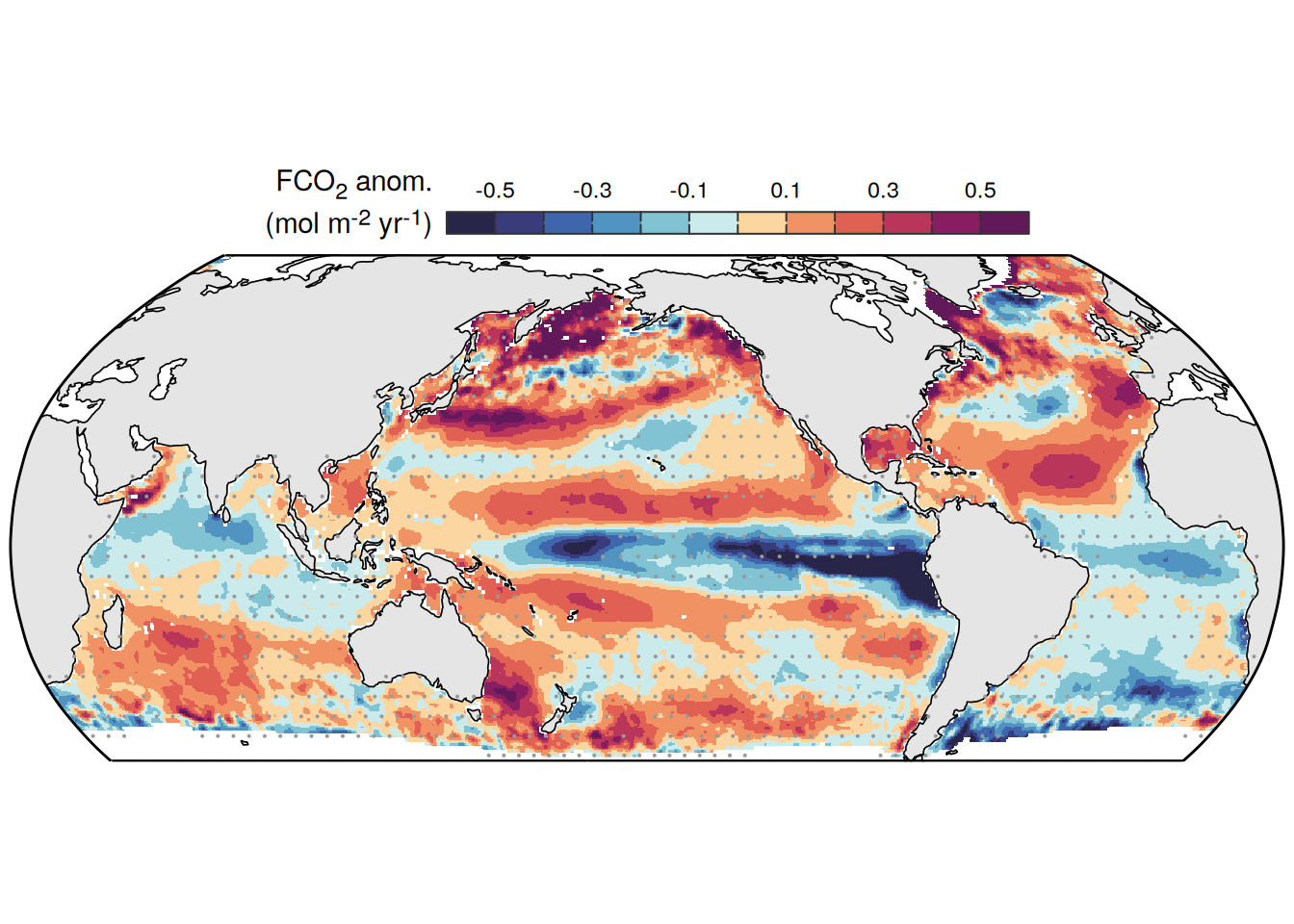
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
[[2]]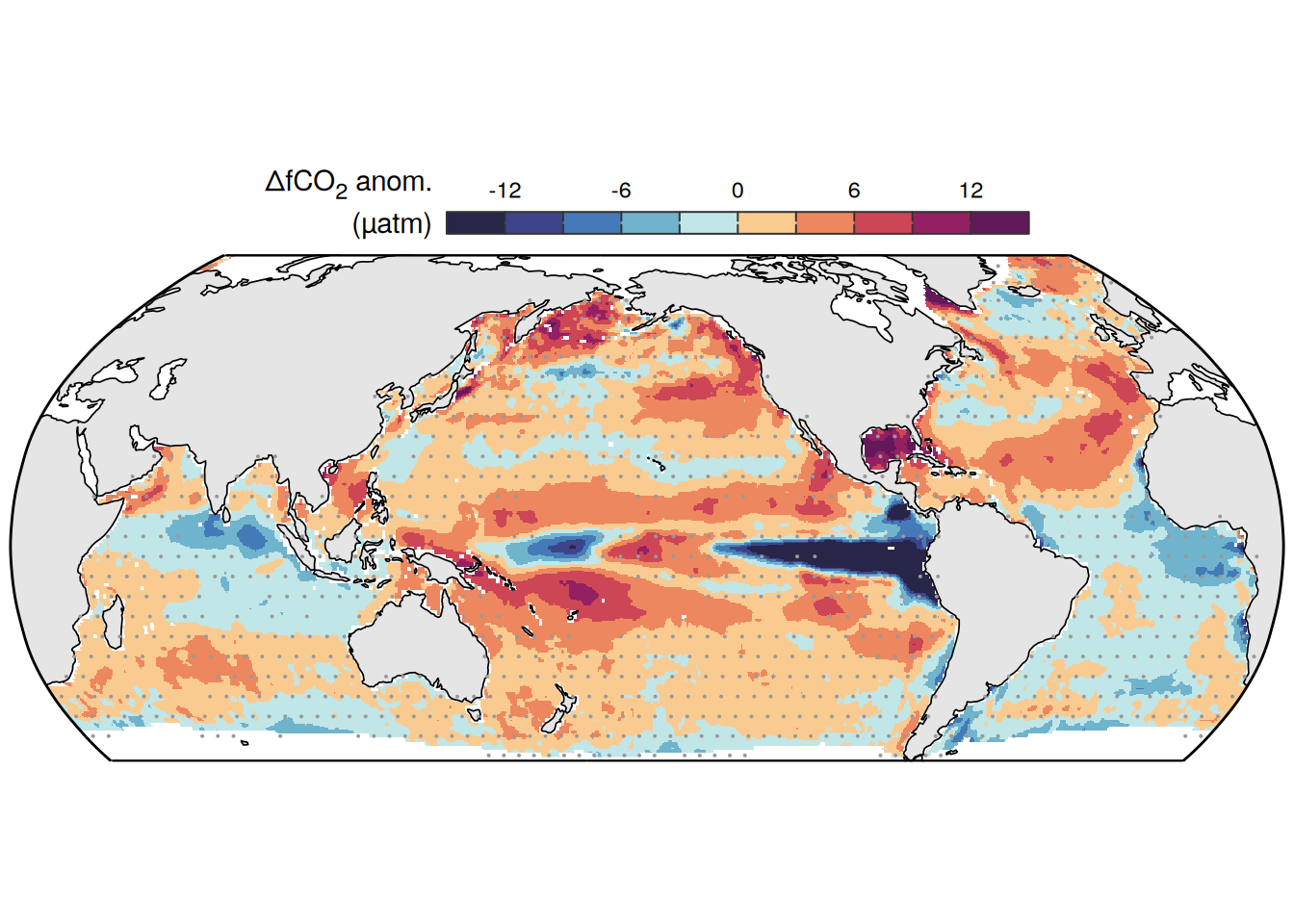
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
[[3]]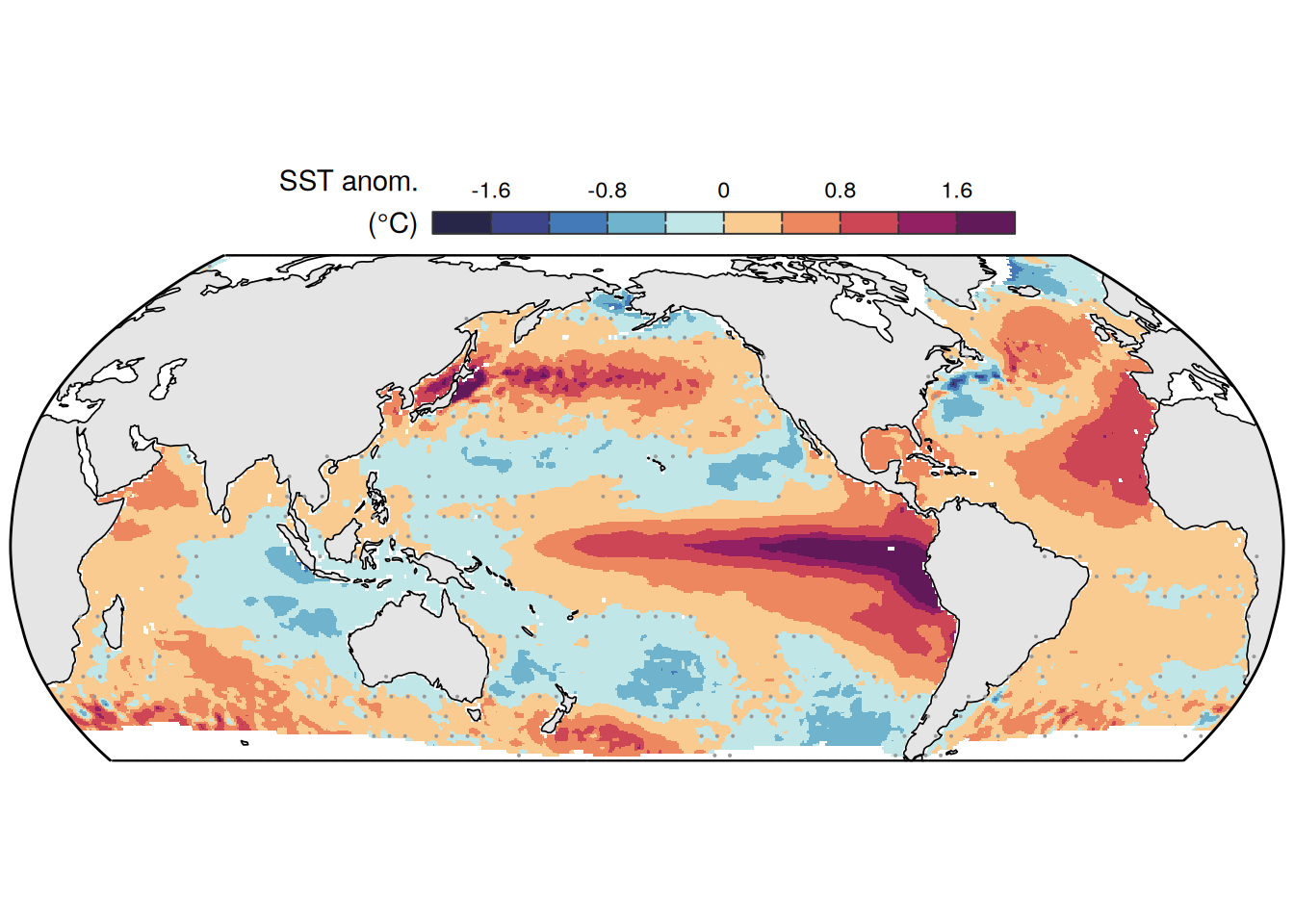
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
[[4]]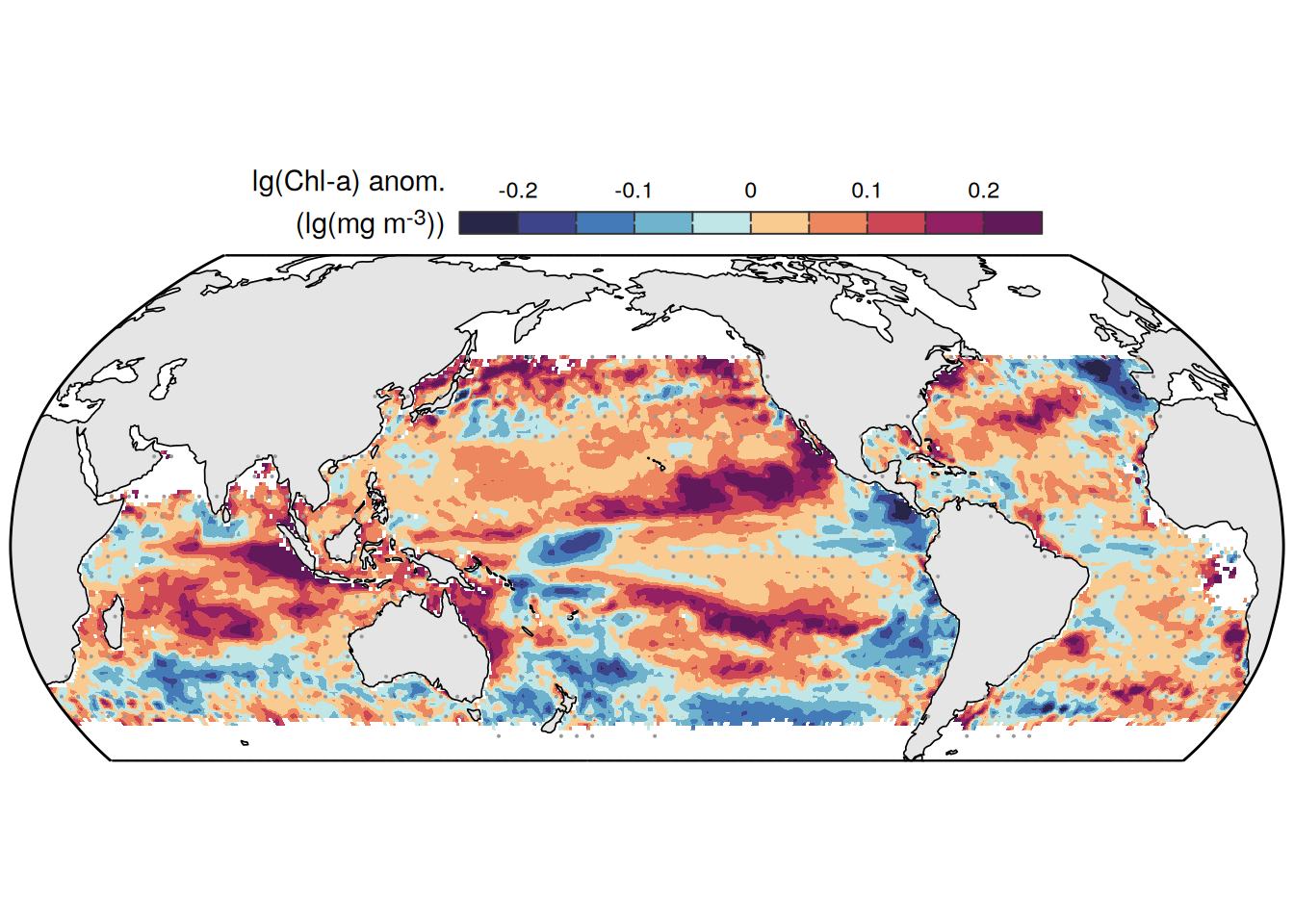
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
plot_list <- pco2_product_map_annual_anomaly_ensemble %>%
mutate(product = "Ensemble mean") %>%
filter(name %in% c("fgco2", "temperature")) %>%
group_split(name) %>%
map(
~ p_map_mdim_robinson(
df = .x,
df_uncertainty = pco2_product_map_annual_anomaly_ensemble_uncertainty %>%
filter(name == .x %>% distinct(name) %>% pull()),
var = "resid_mean",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
legend_position = "bottom",
breaks = labels_breaks(.x %>% distinct(name))$i_breaks,
n_labels = 2
)
)
ggsave(plot = wrap_plots(plot_list,
ncol = 2,
byrow = FALSE),
width = 10,
height = 3,
filename = "../output/map_anomaly_ensemble_mean_pco2_products.jpg")
pco2_product_map_annual_anomaly_ensemble_uncertainty <-
pco2_product_map_annual_anomaly_ensemble_coarse %>%
mutate(signif_single = if_else(abs(value_mean_coarse) < value_sd_coarse, 0, 1)) %>%
select(lon, lat, name, signif_single) %>%
st_as_sf(coords = c("lon", "lat"), crs = "+proj=longlat")
pco2_product_map_annual_anomaly_ensemble %>%
mutate(product = "Ensemble mean") %>%
filter(name %in% c("fgco2")) %>%
group_split(name) %>%
map(
~ p_map_mdim_robinson(
df = .x,
df_uncertainty = pco2_product_map_annual_anomaly_ensemble_uncertainty %>%
filter(name == .x %>% distinct(name) %>% pull()),
var = "value_mean",
legend_title = str_remove(
labels_breaks(.x %>% distinct(name))$i_legend_title,
" anom."),
breaks = c(-Inf, seq(-4,4,1), Inf),
n_labels = 2
)
)[[1]]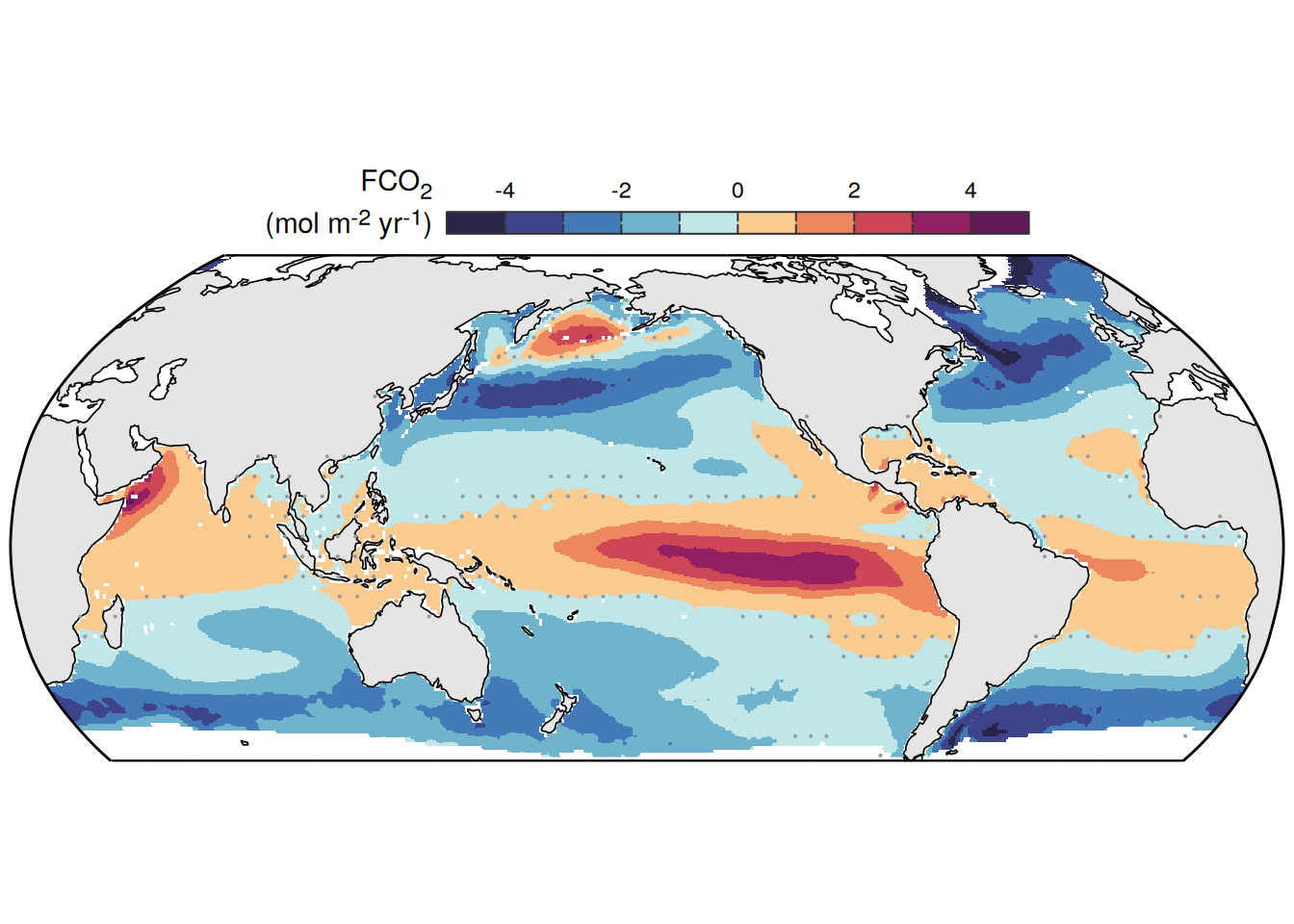
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
ggsave(width = 5,
height = 3,
filename = "../output/map_absolute_ensemble_mean_pco2_products.jpg")
rm(pco2_product_map_annual_anomaly_ensemble_uncertainty)pco2_product_map_annual_anomaly_ensemble_offset <-
left_join(
pco2_product_map_annual_anomaly_ensemble,
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
product %in% pco2_product_list)
) %>%
mutate(`Anomaly offset` = resid - resid_mean) %>%
select(name, lon, lat, product, `Anomaly offset`)
pco2_product_map_annual_anomaly_ensemble_baseline <-
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
product %in% pco2_product_list) %>%
group_by(name, lon, lat) %>%
summarize(
fit_mean = mean(fit),
n = n()
) %>%
ungroup() %>%
filter(n == length(pco2_product_list)) %>%
select(-n)
pco2_product_map_annual_anomaly_ensemble_baseline <-
left_join(
pco2_product_map_annual_anomaly_ensemble_baseline,
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
product %in% pco2_product_list)
) %>%
mutate(`Baseline offset` = fit - fit_mean) %>%
select(name, lon, lat, product, `Baseline offset`)
full_join(
pco2_product_map_annual_anomaly_ensemble_offset,
pco2_product_map_annual_anomaly_ensemble_baseline
) %>%
pivot_longer(contains("offset"),
names_to = "offset") %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = value)) +
labs(title = paste(2023, "offset from ensemble mean")) +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name))$i_legend_title,
limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
oob = squish
) +
facet_grid(product ~ offset) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")
)
rm(pco2_product_map_annual_anomaly_ensemble_offset,
pco2_product_map_annual_anomaly_ensemble_baseline)
gc()pco2_product_map_annual_anomaly_ensemble_gobm <-
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
product %in% gobm_product_list) %>%
group_by(name, lon, lat) %>%
summarize(
resid_sd = sd(resid),
resid_range = max(resid) - min(resid),
resid_mean = mean(resid),
n = n()
) %>%
ungroup() %>%
filter(n == length(gobm_product_list)) %>%
select(-n)
plot_list <-
pco2_product_map_annual_anomaly_ensemble_gobm %>%
filter(name %in% c(
"fgco2",
"dfco2",
"kw_sol",
"temperature",
"salinity",
"sdissic",
"stalk",
"sdissic_stalk",
"no3",
"mld",
"intpp",
"chl"
)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ p_map_mdim_robinson(
df = .x,
var = "resid_mean",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
breaks = labels_breaks(.x %>% distinct(name))$i_breaks
)
)
ggsave(plot = wrap_plots(plot_list,
ncol = 2,
byrow = FALSE),
width = 10,
height = 16,
filename = "../output/map_anomaly_ensemble_mean_gobm.jpg")
rm(plot_list,
pco2_product_map_annual_anomaly_ensemble_gobm)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 3169348 169.3 6540298 349.3 6540298 349.3
Vcells 293256034 2237.4 602973213 4600.4 602973213 4600.4Bivariate anomaly
bivariate_map <-
pco2_product_map_annual_anomaly %>%
filter(year == 2023, name %in% c("fgco2", "temperature")) %>%
select(product, name, lon, lat, resid) %>%
pivot_wider(names_from = name, values_from = resid) %>%
drop_na()
dim_set <- 3
bivariate_map <-
bivariate_map %>%
mutate(
temperature = cut(
temperature,
breaks = c(
min(bivariate_map$temperature),
0,
0.3,
max(bivariate_map$temperature)
),
include.lowest = TRUE
),
fgco2 = cut(
fgco2,
breaks = c(
min(bivariate_map$fgco2),
0,
0.1,
max(bivariate_map$fgco2)
),
include.lowest = TRUE
)
)
bivariate_map <-
bi_class(
bivariate_map,
x = temperature,
y = fgco2,
dim = dim_set,
style = "quantile"
)
bi_breaks <-
bi_class_breaks(
bivariate_map,
x = temperature,
y = fgco2,
dim = dim_set,
style = "quantile",
dig_lab = 1,
split = TRUE
)
bivariate_map_raster <-
bivariate_map %>%
relocate(lon, lat) %>%
select(lon, lat, product, bi_class) %>%
mutate(bi_class_numeric = as.character(as.numeric(as.factor(bi_class))))
bivariate_map_raster_values <-
bivariate_map_raster %>%
distinct(bi_class, bi_class_numeric)
bivariate_map_raster <- rast(
bivariate_map_raster %>%
select(-bi_class) %>%
pivot_wider(names_from = product,
values_from = bi_class_numeric),
crs = "+proj=longlat"
)
bivariate_map_raster <- project(bivariate_map_raster, target_crs, method = "near")
bivariate_map_tibble <- bivariate_map_raster %>%
as.data.frame(xy = TRUE, na.rm = FALSE) %>%
as_tibble() %>%
rename(lon = x, lat = y) %>%
pivot_longer(-c(lon, lat),
names_to = "product",
values_to = "bi_class_numeric") %>%
drop_na()
bivariate_map_tibble <-
right_join(
bivariate_map_tibble,
bivariate_map_raster_values %>%
mutate(bi_class_numeric = as.numeric(bi_class_numeric))
)
ggplot() +
geom_raster(data = bivariate_map_tibble,
aes(x = lon, y = lat, fill = bi_class)) +
bi_scale_fill(pal = "DkBlue2", dim = dim_set, flip_axes = TRUE) +
geom_sf(data = worldmap_trans, fill = "grey90", col = "grey90") +
geom_sf(data = coastline_trans, linewidth = 0.3) +
geom_sf(data = bbox_graticules_trans, linewidth = 0.5) +
coord_sf(
crs = target_crs,
ylim = lat_lim,
xlim = lon_lim,
expand = FALSE
) +
theme(
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank(),
panel.border = element_rect(colour = "transparent"),
strip.background = element_blank(),
legend.position = "none"
) +
facet_wrap( ~ product, ncol = 2)
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 681629f | jens-daniel-mueller | 2024-08-26 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
ggsave(
width = 6,
height = 5,
dpi = 600,
filename = "../output/map_anomaly_bivariate_all_products.jpg"
)
bi_breaks$bi_x <- bi_breaks$bi_x[-1]
bi_breaks$bi_x[1] <- paste0("-", bi_breaks$bi_x[1])
bi_breaks$bi_y <- bi_breaks$bi_y[-1]
bi_breaks$bi_y[1] <- paste0("-", bi_breaks$bi_y[1])
bi_legend(
pal = "DkBlue2",
xlab = labels_breaks("temperature")$i_legend_title,
ylab = labels_breaks("fgco2")$i_legend_title,
dim = dim_set,
pad_width = 2,
breaks = bi_breaks,
arrows = FALSE,
flip_axes = TRUE
) +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
axis.ticks = element_blank(),
axis.text = element_text(size = 10)
)
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 681629f | jens-daniel-mueller | 2024-08-26 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 960912c | jens-daniel-mueller | 2024-05-16 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
ggsave(
width = 4,
height = 3,
dpi = 600,
filename = "../output/map_anomaly_bivariate_all_products_legend.jpg"
)bivariate_map <-
pco2_product_map_annual_anomaly_ensemble %>%
filter(name %in% c("fgco2", "temperature")) %>%
select(name, lon, lat, resid_mean) %>%
pivot_wider(names_from = name,
values_from = resid_mean) %>%
drop_na()
dim_set <- 3
bivariate_map <-
bivariate_map %>%
mutate(
temperature = cut(
temperature,
breaks = c(
min(bivariate_map$temperature),
0,
0.3,
max(bivariate_map$temperature)
),
include.lowest = TRUE
),
fgco2 = cut(
fgco2,
breaks = c(
max(bivariate_map$fgco2),
0.1,
0,
min(bivariate_map$fgco2)
),
include.lowest = TRUE
)
)
bivariate_map <-
bi_class(
bivariate_map,
x = temperature,
y = fgco2,
dim = dim_set,
style = "quantile"
)
bi_breaks <-
bi_class_breaks(
bivariate_map,
x = temperature,
y = fgco2,
dim = dim_set,
style = "quantile",
dig_lab = 1,
split = TRUE
)
bivariate_map_raster <-
bivariate_map %>%
relocate(lon, lat) %>%
select(lon, lat, bi_class) %>%
mutate(bi_class_numeric = as.character(as.numeric(as.factor(bi_class))))
bivariate_map_raster_values <-
bivariate_map_raster %>%
distinct(bi_class, bi_class_numeric)
bivariate_map_raster <- rast(
bivariate_map_raster %>%
select(-bi_class),
crs = "+proj=longlat"
)
bivariate_map_raster <- project(bivariate_map_raster, target_crs, method = "near")
bivariate_map_tibble <- bivariate_map_raster %>%
as.data.frame(xy = TRUE, na.rm = FALSE) %>%
as_tibble() %>%
rename(lon = x, lat = y) %>%
drop_na()
bivariate_map_tibble <-
right_join(
bivariate_map_tibble,
bivariate_map_raster_values %>%
mutate(bi_class_numeric = as.numeric(bi_class_numeric))
)
ggplot() +
geom_raster(data = bivariate_map_tibble,
aes(x = lon, y = lat, fill = bi_class)) +
bi_scale_fill(pal = "DkBlue2", dim = dim_set, flip_axes = TRUE) +
geom_sf(data = worldmap_trans, fill = "grey90", col = "grey90") +
geom_sf(data = coastline_trans, linewidth = 0.3) +
geom_sf(data = bbox_graticules_trans, linewidth = 0.5) +
coord_sf(
crs = target_crs,
ylim = lat_lim,
xlim = lon_lim,
expand = FALSE
) +
theme(
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank(),
panel.border = element_rect(colour = "transparent"),
strip.background = element_blank(),
legend.position = "none"
)
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 681629f | jens-daniel-mueller | 2024-08-26 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
ggsave(width = 5,
height = 2.5,
dpi = 600,
filename = "../output/map_anomaly_bivariate_ensemble_mean_pco2_products.jpg")
bi_breaks$bi_x <- bi_breaks$bi_x[-1]
bi_breaks$bi_x[1] <- paste0("-", bi_breaks$bi_x[1])
bi_breaks$bi_y <- bi_breaks$bi_y[-1]
bi_breaks$bi_y[1] <- paste0("-", bi_breaks$bi_y[1])
bi_legend(
pal = "DkBlue2",
xlab = labels_breaks("temperature")$i_legend_title,
ylab = labels_breaks("fgco2")$i_legend_title,
dim = dim_set,
pad_width = 2,
breaks = bi_breaks,
arrows = FALSE,
flip_axes = TRUE
) +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
axis.ticks = element_blank(),
axis.text = element_text(size = 10)
)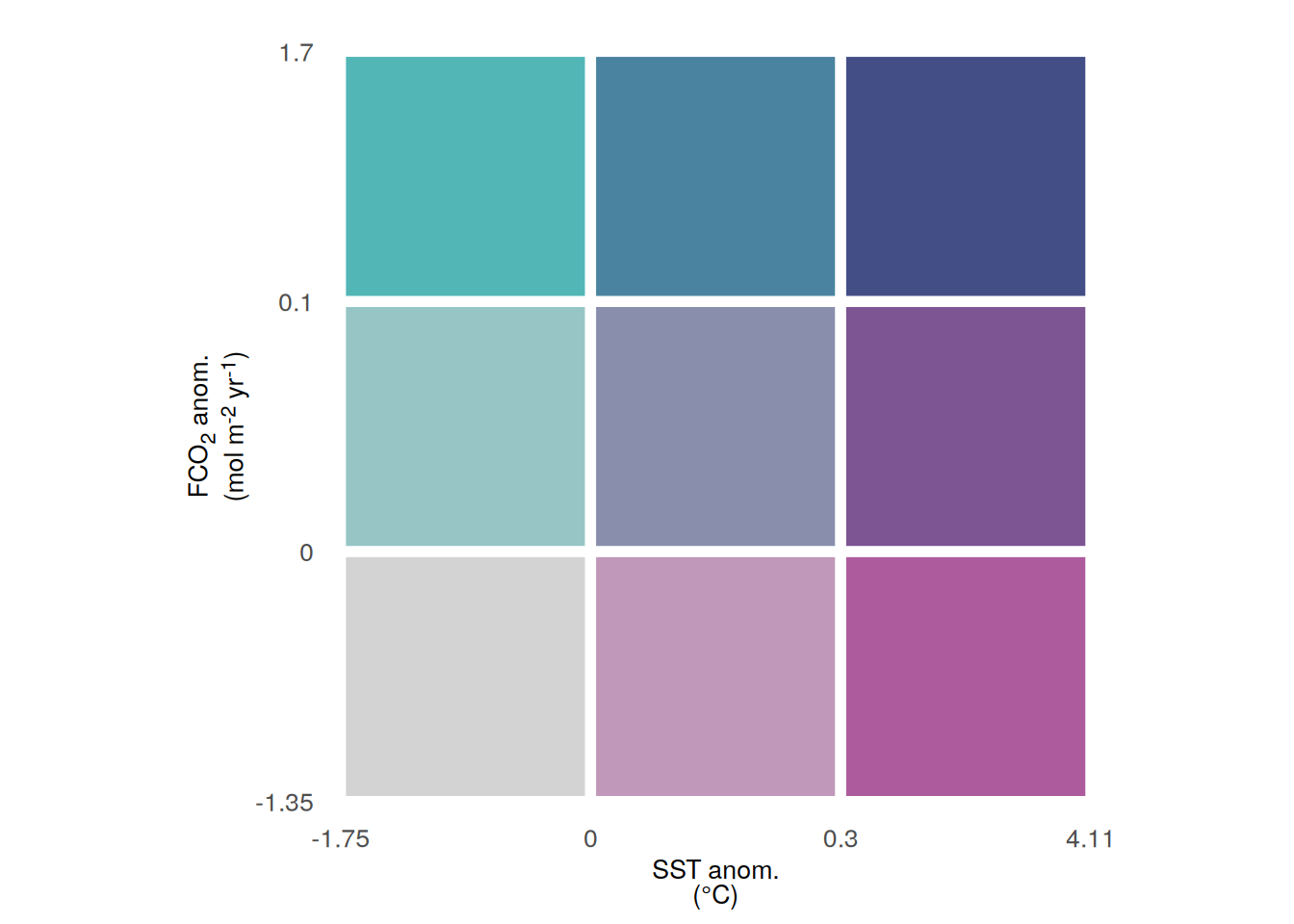
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 681629f | jens-daniel-mueller | 2024-08-26 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| aeca619 | jens-daniel-mueller | 2024-06-19 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
ggsave(width = 4,
height = 3,
dpi = 600,
filename = "../output/map_anomaly_bivariate_ensemble_mean_pco2_products_legend.jpg")pco2_product_zonal_annual_anomaly <-
pco2_product_hovmoeller_monthly_anomaly %>%
filter(year == 2023) %>%
group_by(product, name, lat) %>%
summarise(resid = mean(resid)) %>%
ungroup()
pco2_product_zonal_annual_anomaly %>%
ggplot(aes(resid, lat, col = product)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 0) +
geom_path() +
scale_color_manual(values = color_products) +
facet_wrap( ~ name, scales = "free_x", ncol = 4)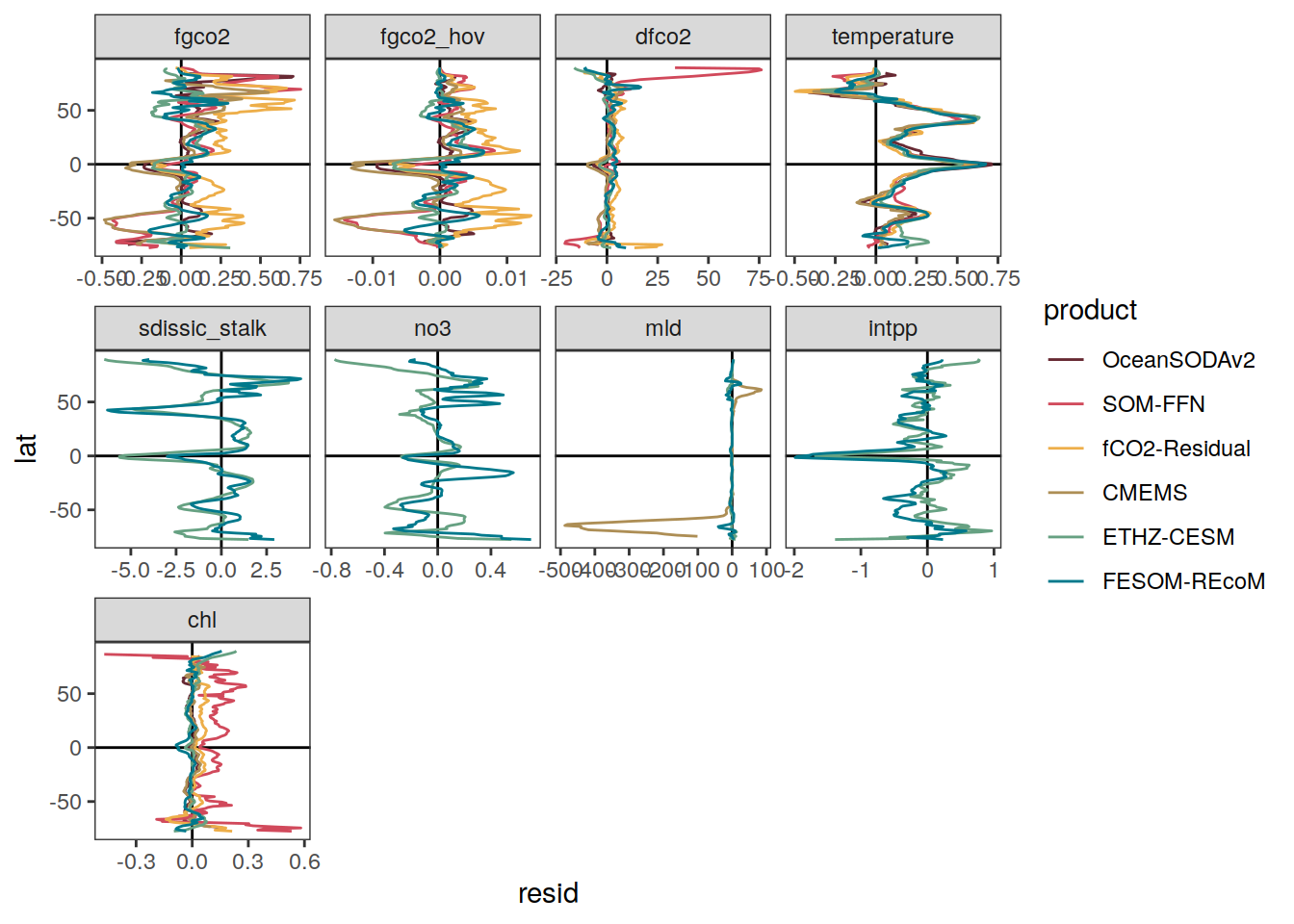
pco2_product_zonal_annual_anomaly_ensemble <-
pco2_product_zonal_annual_anomaly %>%
filter(product %in% pco2_product_list) %>%
group_by(lat, name) %>%
fsummarise(
resid_sd = fsd(resid),
resid_mean = fmean(resid)
)
pco2_product_zonal_annual_anomaly_ensemble %>%
filter(name %in% c("fgco2_hov", "temperature")) %>%
ggplot(aes(resid_mean, lat)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 0) +
# geom_ribbon(aes(xmin = resid_mean - resid_sd, xmax = resid_mean + resid_sd),
# alpha = 0.5) +
geom_ribbon(aes(xmin = 0, xmax = pmax(0, resid_mean), fill = "Positive"),
alpha = 0.5) +
geom_ribbon(aes(xmax = 0, xmin = pmin(0, resid_mean), fill = "Negative"),
alpha = 0.5) +
scale_fill_manual(values = c(cold_color, warm_color)) +
geom_path() +
facet_grid(. ~ name,
labeller = labeller(name = x_axis_labels),
scales = "free_x",
switch = "x") +
scale_y_continuous(breaks = seq(-60,60,30),
name = "Lat (°N)",
limits = c(-54,76),
expand = c(0,0)) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.x = element_blank(),
legend.position = "none"
)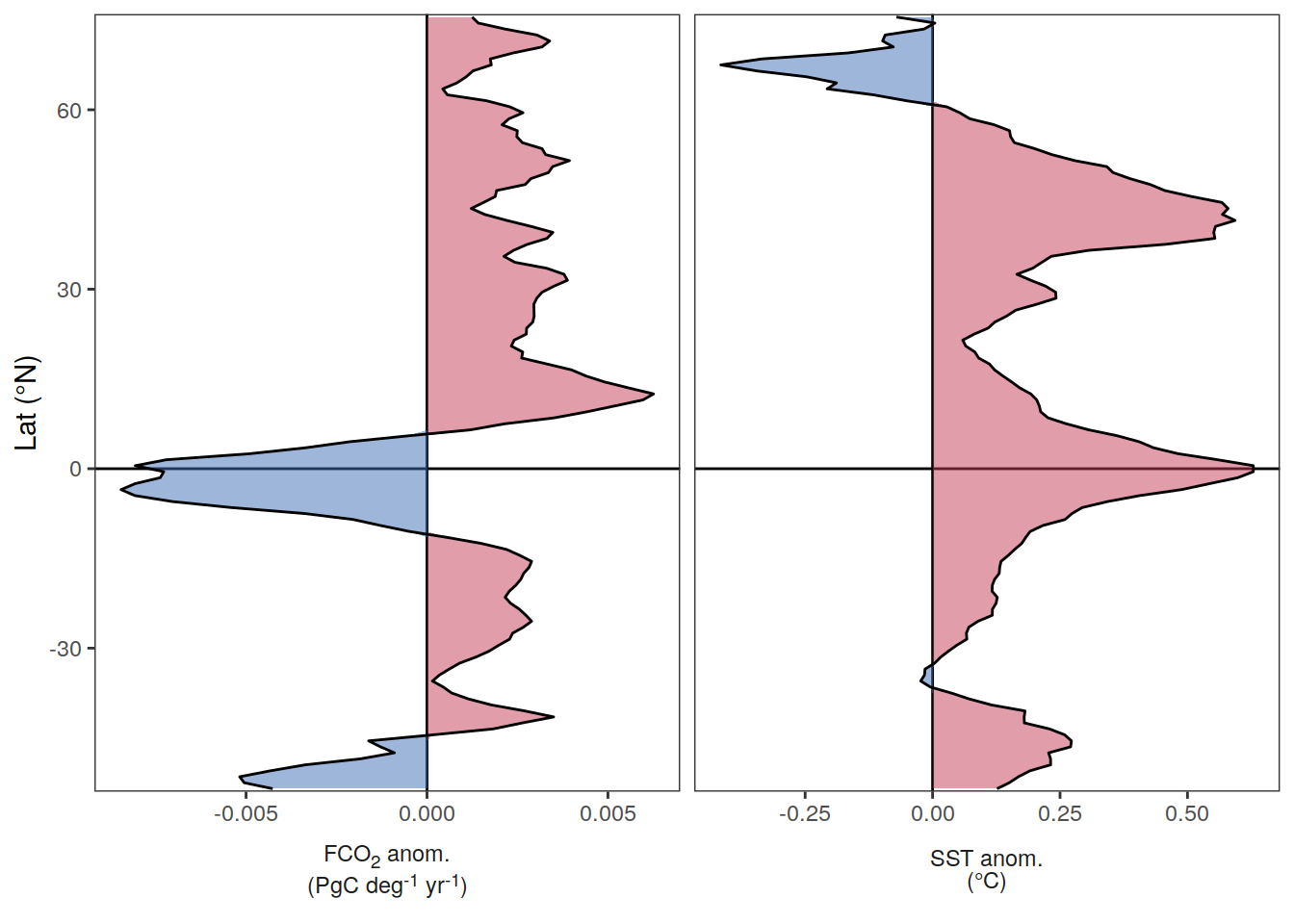
bi_pal("DkBlue2", preview = FALSE) 1-1 2-1 3-1 1-2 2-2 3-2 1-3 2-3
"#d3d3d3" "#97c5c5" "#52b6b6" "#c098b9" "#898ead" "#4a839f" "#ad5b9c" "#7c5592"
3-3
"#434e87" # "#d3d3d3" "#97c5c5" "#52b6b6" "#c098b9" "#898ead" "#4a839f" "#ad5b9c" "#7c5592" "#434e87"
p_zonal_fgco2 <-
pco2_product_zonal_annual_anomaly_ensemble %>%
filter(name %in% c("fgco2_hov")) %>%
mutate(resid_mean = resid_mean * 1000) %>%
ggplot(aes(resid_mean, lat)) +
geom_vline(xintercept = 0) +
geom_ribbon(aes(xmin = 0, xmax = pmax(0, resid_mean), fill = "Positive"),
alpha = 0.9) +
geom_ribbon(aes(xmax = 0, xmin = pmin(0, resid_mean), fill = "Negative"),
alpha = 0.9) +
scale_fill_manual(values = c("#d3d3d3", "#52b6b6")) +
geom_path() +
scale_y_continuous(breaks = seq(-60,60,30),
name = "Lat (°N)",
limits = c(-54,76),
expand = c(0,0)) +
scale_x_continuous(breaks = seq(-5,5,5),
name = str_replace(
labels_breaks("fgco2_hov")$i_legend_title,
"PgC", "TgC"
)) +
theme_classic() +
theme(
legend.position = "none",
axis.title.x = element_markdown(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.line.y = element_blank()
)
p_zonal_temperature <-
pco2_product_zonal_annual_anomaly_ensemble %>%
filter(name %in% c("temperature")) %>%
ggplot(aes(resid_mean, lat)) +
geom_vline(xintercept = 0) +
geom_ribbon(aes(xmin = 0, xmax = pmax(0, resid_mean), fill = "Positive"),
alpha = 0.9) +
geom_ribbon(aes(xmax = 0, xmin = pmin(0, resid_mean), fill = "Negative"),
alpha = 0.9) +
scale_fill_manual(values = c("#d3d3d3", "#ad5b9c")) +
geom_path() +
scale_y_continuous(breaks = seq(-60,60,30),
name = "Lat (°N)",
limits = c(-54,76),
expand = c(0,0)) +
scale_x_continuous(breaks = seq(-0.6,0.6,0.3),
name = labels_breaks("temperature")$i_legend_title) +
theme_classic() +
theme(
legend.position = "none",
axis.title.x = element_markdown(),
axis.title.y = element_text(angle = 0)
)
p_zonal_temperature | p_zonal_fgco2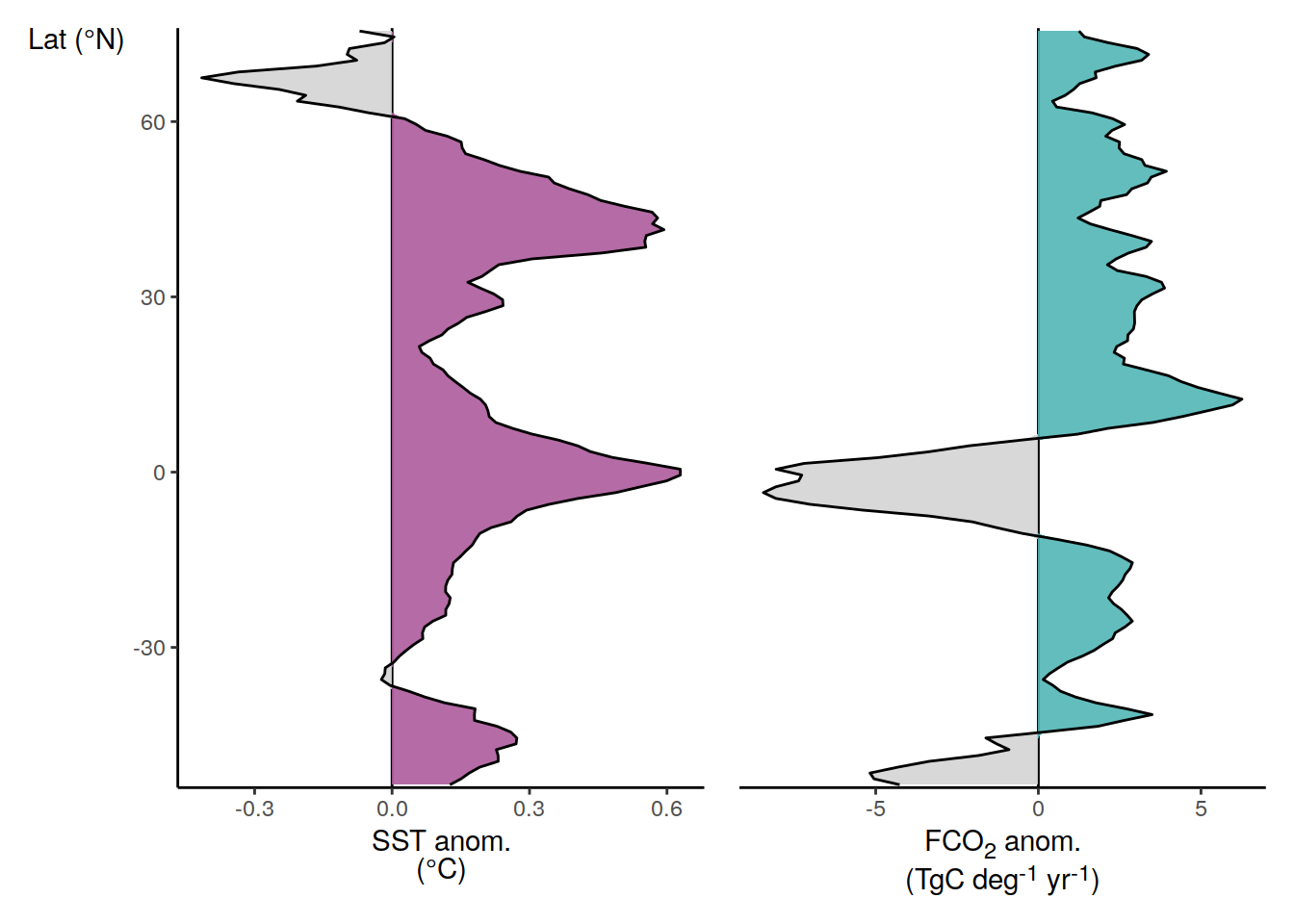
ggsave(width = 2.8,
height = 4.5,
filename = "../output/zonal_mean_anomaly_pco2_product_ensemble_mean_bivariate_color.jpg")
p_zonal_fgco2 <-
pco2_product_zonal_annual_anomaly_ensemble %>%
filter(name %in% c("fgco2_hov")) %>%
mutate(resid_mean = resid_mean * 1000) %>%
ggplot(aes(resid_mean, lat)) +
geom_vline(xintercept = 0) +
geom_ribbon(aes(xmin = 0, xmax = pmax(0, resid_mean), fill = "Positive"),
alpha = 0.9) +
geom_ribbon(aes(xmax = 0, xmin = pmin(0, resid_mean), fill = "Negative"),
alpha = 0.9) +
scale_fill_manual(values = c(cold_color, warm_color)) +
geom_path() +
scale_y_continuous(breaks = seq(-60,60,30),
name = "Lat (°N)",
limits = c(-54,76),
expand = c(0,0)) +
scale_x_continuous(breaks = seq(-5,5,5),
name = str_replace(
labels_breaks("fgco2_hov")$i_legend_title,
"PgC", "TgC"
)) +
theme_classic() +
theme(
legend.position = "none",
axis.title.x = element_markdown(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.line.y = element_blank()
)
p_zonal_temperature <-
pco2_product_zonal_annual_anomaly_ensemble %>%
filter(name %in% c("temperature")) %>%
ggplot(aes(resid_mean, lat)) +
geom_vline(xintercept = 0) +
geom_ribbon(aes(xmin = 0, xmax = pmax(0, resid_mean), fill = "Positive"),
alpha = 0.9) +
geom_ribbon(aes(xmax = 0, xmin = pmin(0, resid_mean), fill = "Negative"),
alpha = 0.9) +
scale_fill_manual(values = c(cold_color, warm_color)) +
geom_path() +
scale_y_continuous(breaks = seq(-60,60,30),
name = "Lat (°N)",
limits = c(-54,76),
expand = c(0,0)) +
scale_x_continuous(breaks = seq(-0.6,0.6,0.3),
name = labels_breaks("temperature")$i_legend_title) +
theme_classic() +
theme(
legend.position = "none",
axis.title.x = element_markdown(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.line.y = element_blank()
)
p_zonal_temperature | p_zonal_fgco2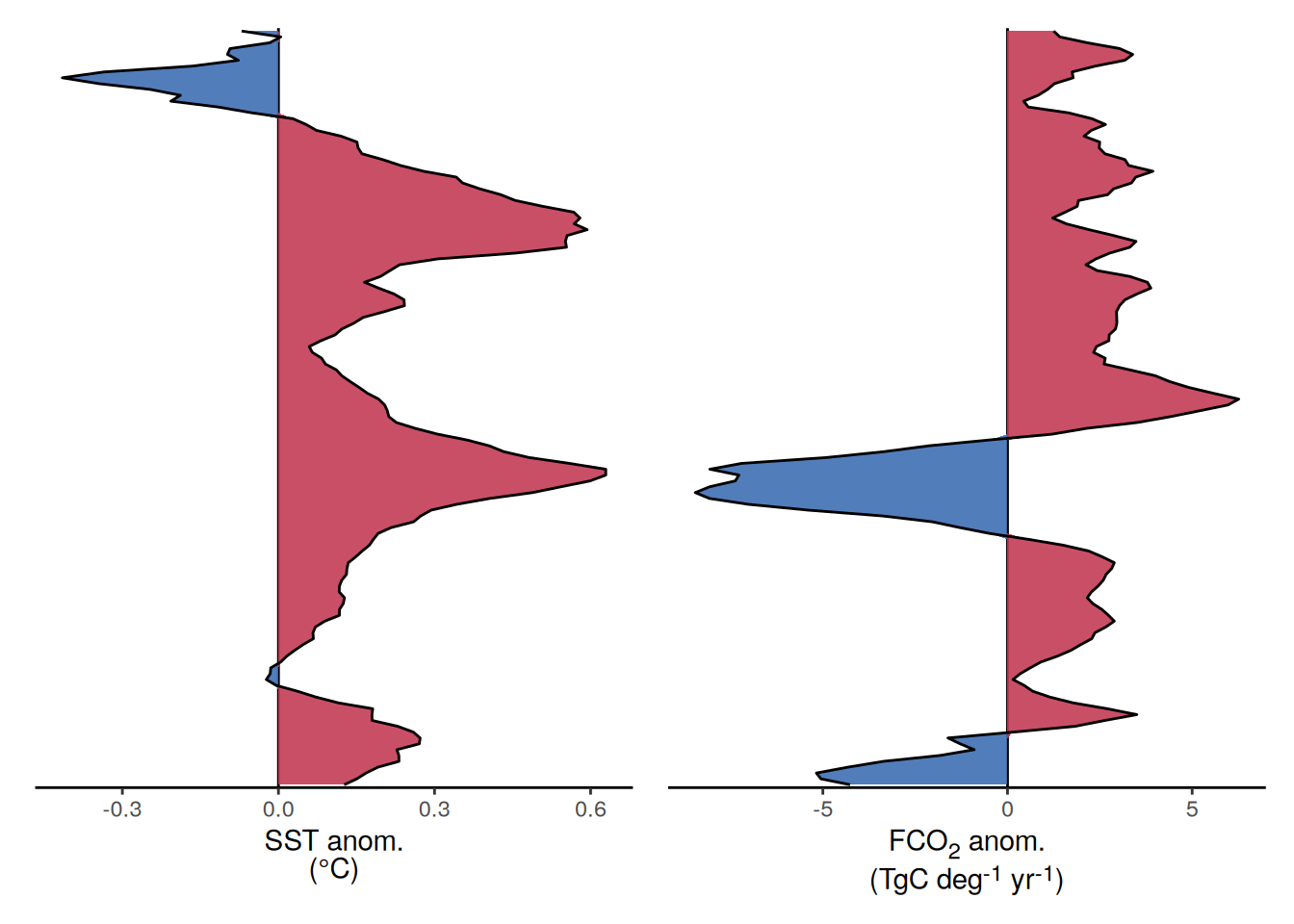
ggsave(width = 2.5,
height = 4,
filename = "../output/zonal_mean_anomaly_pco2_product_ensemble_mean_warm_cold_color.jpg")SST flux slope
pco2_product_map_annual_anomaly_temperature_predict <-
pco2_product_map_annual_anomaly_temperature_predict %>%
drop_na()
pco2_product_map_annual_anomaly_temperature_predict %>%
p_map_mdim_robinson(
var = "slope",
legend_title = "Slope FCO<sub>2</sub> anom. / SST anom.<br>(mol m<sup>-2</sup> yr<sup>-1</sup> °C<sup>-1</sup>)",
breaks = c(-Inf, seq(-1, 1, 0.25), Inf),
dim_wrap = "product",
n_col = 2
)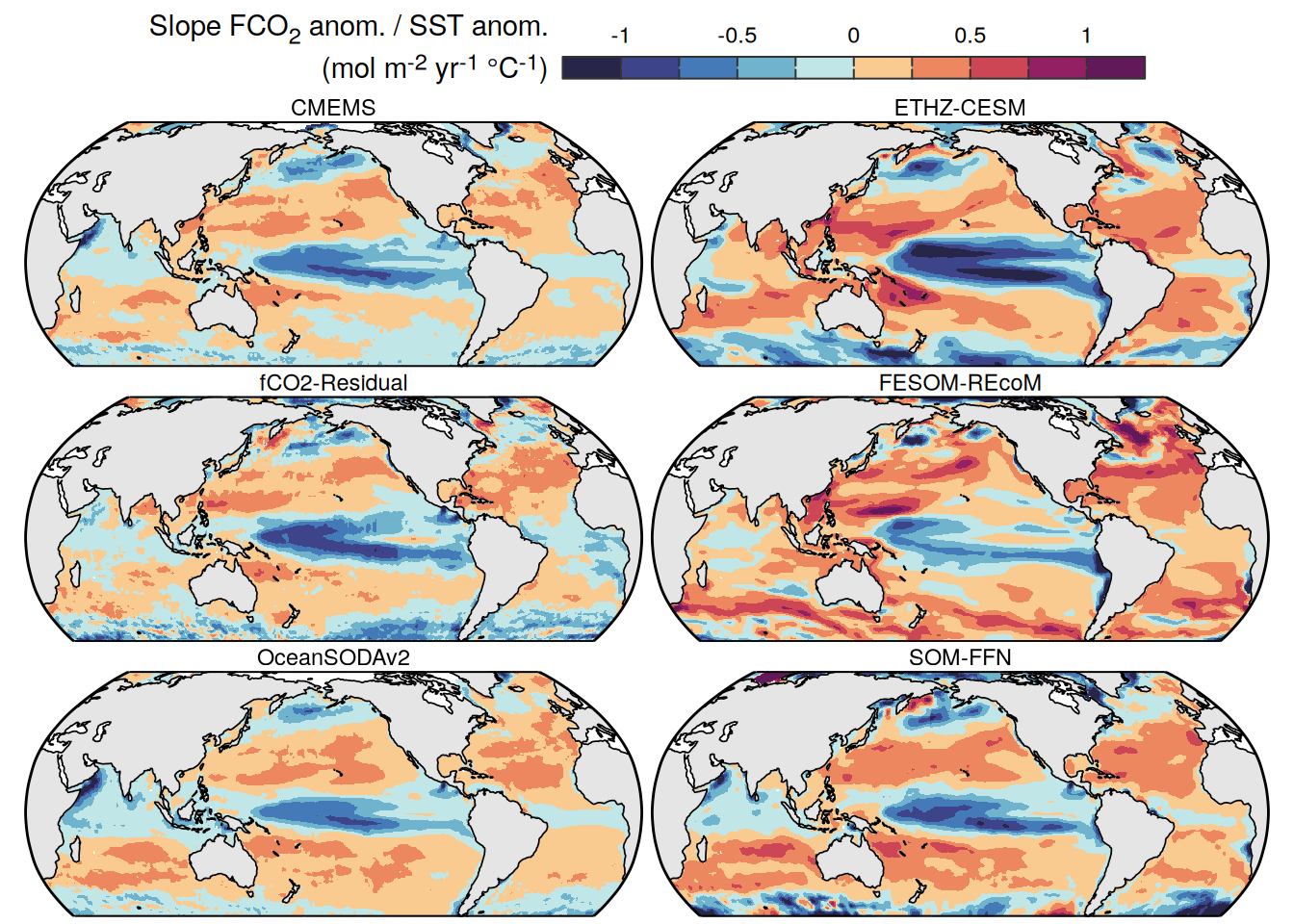
ggsave(width = 7,
height = 6,
dpi = 600,
filename = "../output/map_anomaly_correlation_all_products.jpg")
pco2_product_map_annual_anomaly_temperature_predict <-
pco2_product_map_annual_anomaly_temperature_predict %>%
select(-year) %>%
pivot_longer(-c(product, lon, lat), values_to = "resid")
pco2_product_map_annual_anomaly_temperature_predict %>%
filter(str_detect(name, "fgco2")) %>%
p_map_mdim_robinson(
var = "resid",
legend_title = labels_breaks("fgco2")$i_legend_title,
breaks = labels_breaks("fgco2")$i_breaks,
dim_row = "product",
dim_col = "name"
)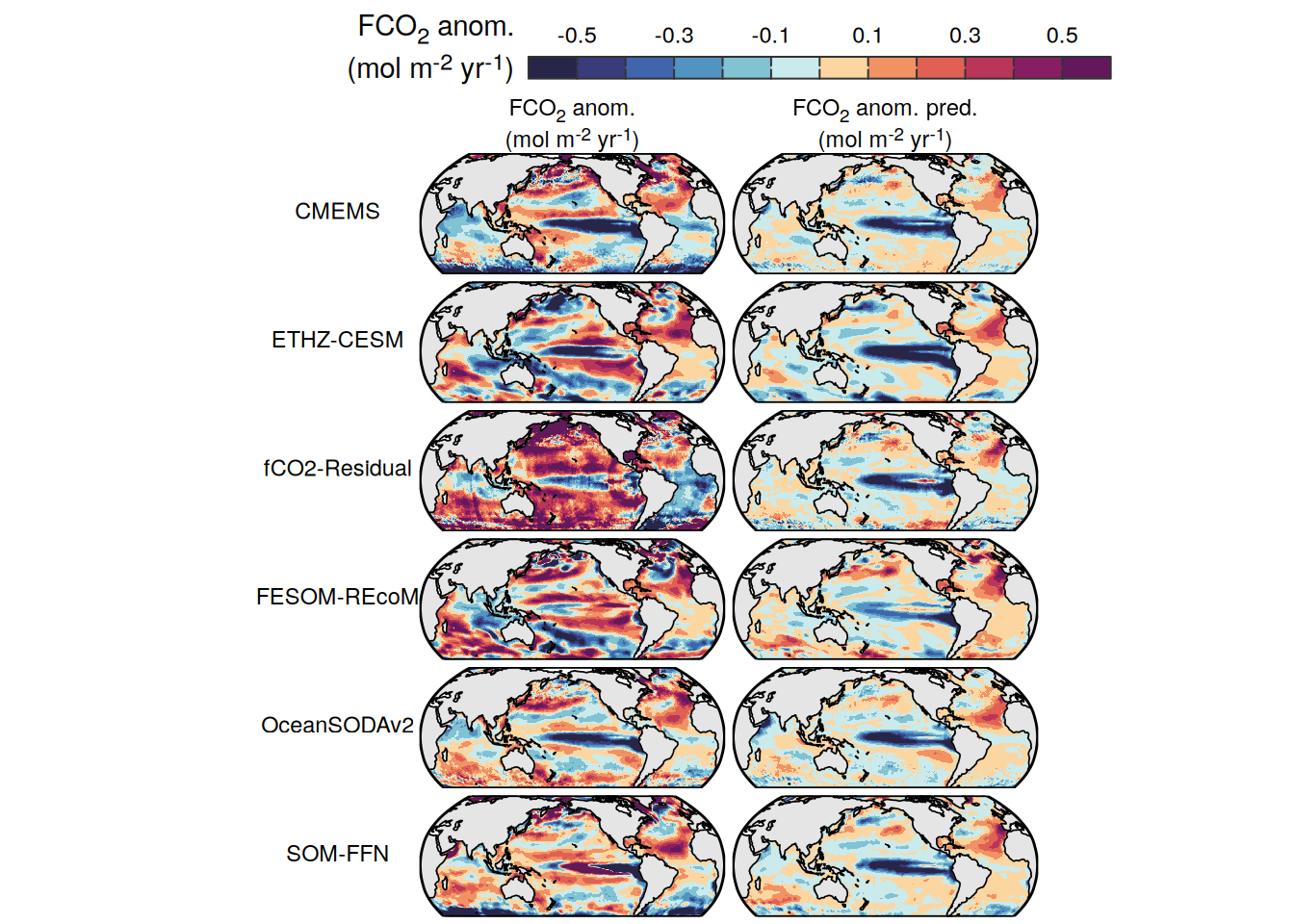
pco2_product_map_annual_anomaly_temperature_predict_ensemble <-
pco2_product_map_annual_anomaly_temperature_predict %>%
filter(product %in% pco2_product_list) %>%
fgroup_by(name, lon, lat) %>%
fsummarise(
resid_sd = fsd(resid),
resid_mean = fmean(resid),
n = fnobs(resid)
) %>%
filter(n == length(pco2_product_list)) %>%
select(-n)
pco2_product_map_annual_anomaly_temperature_predict_ensemble_coarse <-
m_grid_horizontal_coarse(pco2_product_map_annual_anomaly_temperature_predict_ensemble) %>%
fgroup_by(name, lon_grid, lat_grid) %>%
fsummarise(
resid_sd_coarse = fmean(resid_sd, na.rm = TRUE),
resid_mean_coarse = fmean(resid_mean, na.rm = TRUE)
) %>%
rename(lon = lon_grid, lat = lat_grid)
pco2_product_map_annual_anomaly_temperature_predict_ensemble_uncertainty <-
pco2_product_map_annual_anomaly_temperature_predict_ensemble_coarse %>%
mutate(signif_single = if_else(abs(resid_mean_coarse) < resid_sd_coarse, 0, 1)) %>%
select(lon, lat, name, signif_single) %>%
st_as_sf(coords = c("lon", "lat"), crs = "+proj=longlat")
pco2_product_map_annual_anomaly_temperature_predict_ensemble %>%
mutate(product = "Ensemble mean") %>%
group_split(name) %>%
# head(1) %>%
map(
~ p_map_mdim_robinson(
df = .x,
df_uncertainty = pco2_product_map_annual_anomaly_temperature_predict_ensemble_uncertainty %>%
filter(name == .x %>% distinct(name) %>% pull()),
var = "resid_mean",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
breaks = labels_breaks(.x %>% distinct(name))$i_breaks,
n_labels = 2
)
)[[1]]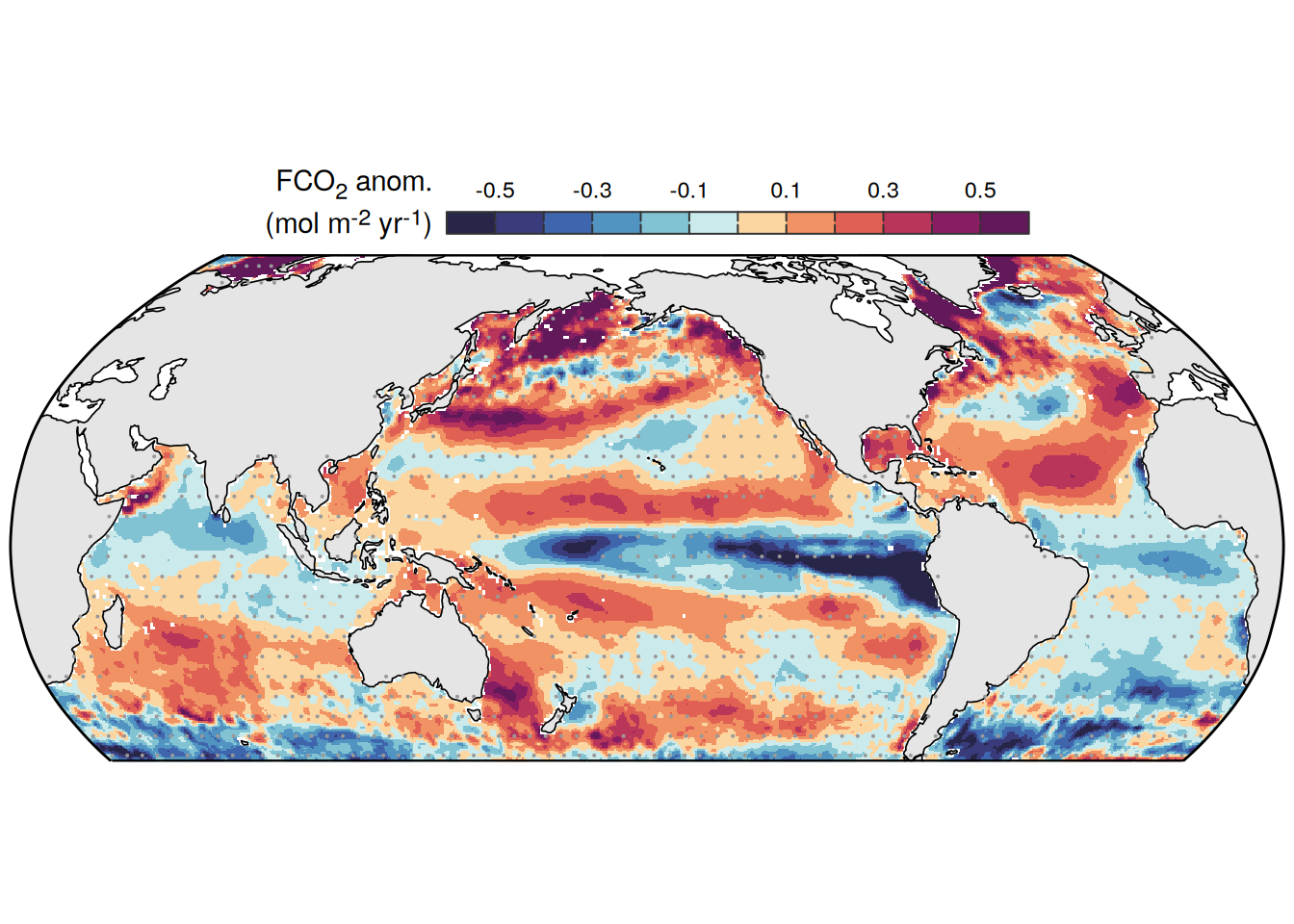
[[2]]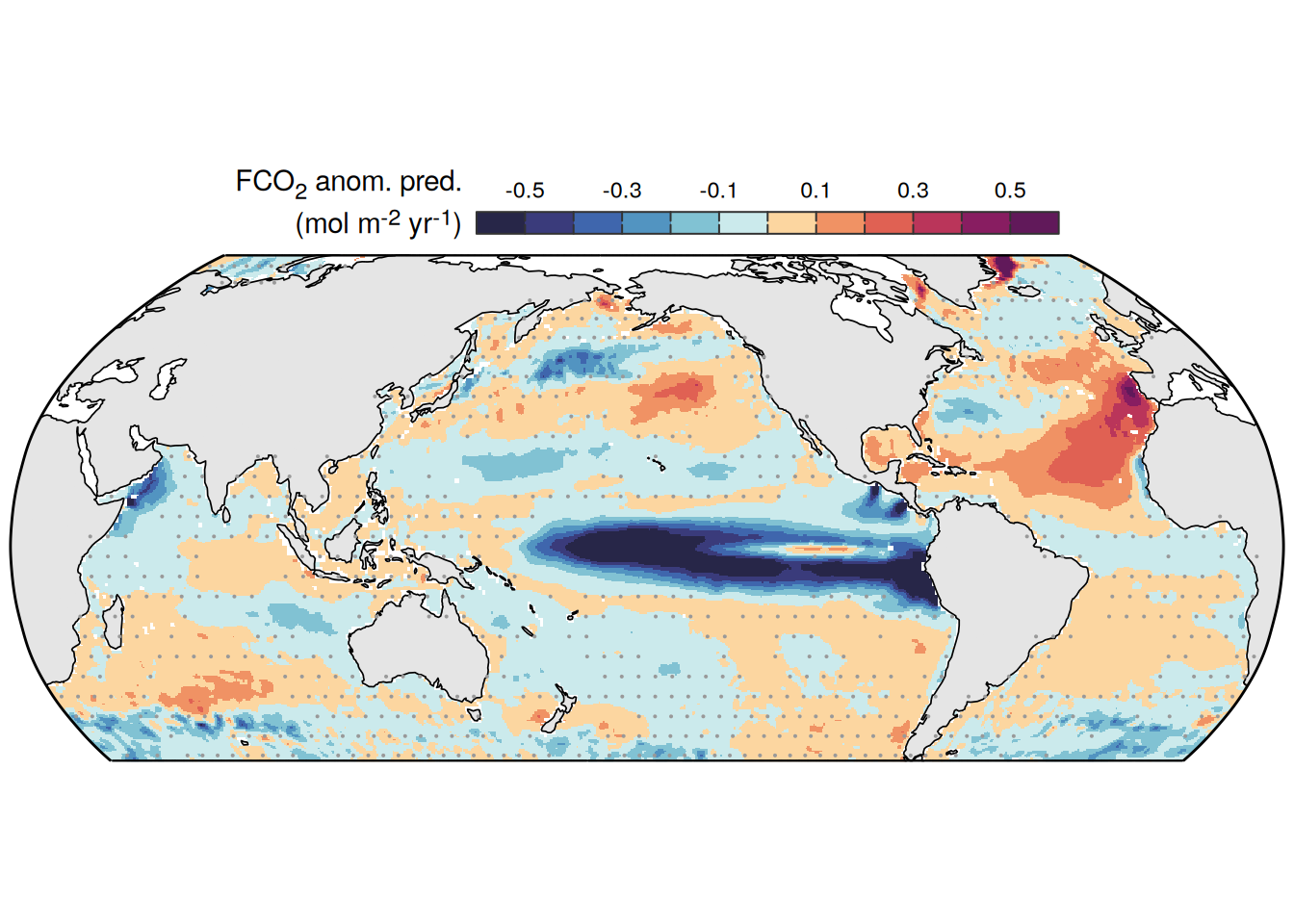
[[3]]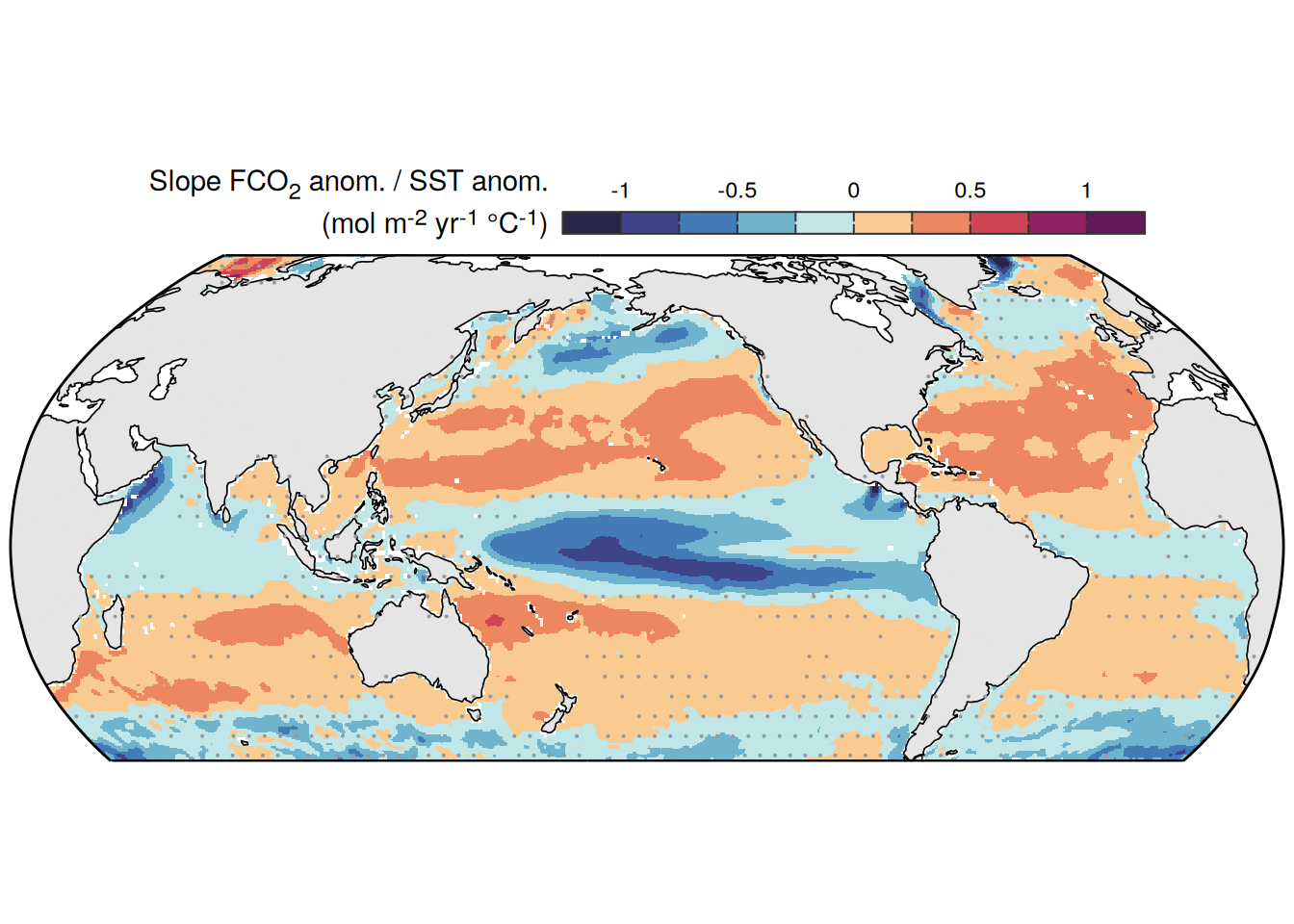
[[4]]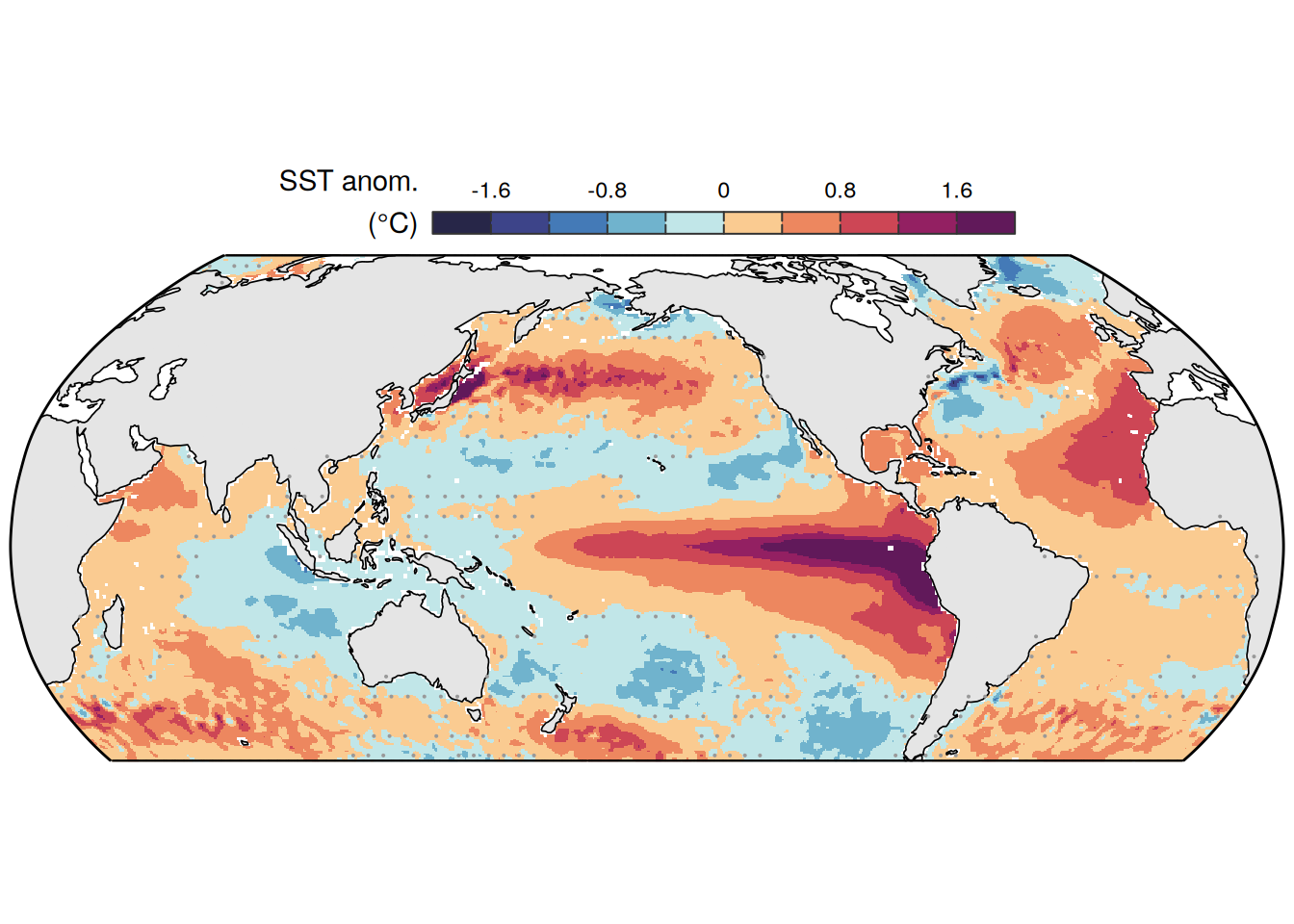
plot_list <- pco2_product_map_annual_anomaly_temperature_predict_ensemble %>%
mutate(product = "Ensemble mean") %>%
group_split(name) %>%
map(
~ p_map_mdim_robinson(
df = .x,
df_uncertainty = pco2_product_map_annual_anomaly_temperature_predict_ensemble_uncertainty %>%
filter(name == .x %>% distinct(name) %>% pull()),
var = "resid_mean",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
legend_position = "bottom",
breaks = labels_breaks(.x %>% distinct(name))$i_breaks,
n_labels = 2
)
)
ggsave(plot = wrap_plots(plot_list,
ncol = 2),
width = 12,
height = 6,
filename = "../output/map_annual_anomaly_temperature_predict_ensemble.jpg")
rm(
pco2_product_map_annual_anomaly_temperature_predict_ensemble,
pco2_product_map_annual_anomaly_temperature_predict_ensemble_coarse,
pco2_product_map_annual_anomaly_temperature_predict_ensemble_uncertainty
)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 3305645 176.6 6540298 349.3 6540298 349.3
Vcells 310945970 2372.4 602973213 4600.4 602973213 4600.4Monthly means
2023 anomaly
pco2_product_map_monthly_anomaly <-
inner_join(
biome_mask_print,
pco2_product_map_monthly_anomaly
)pco2_product_map_monthly_anomaly %>%
filter(name %in% name_core,
year == 2023) %>%
group_split(name) %>%
head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid)) +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name))$i_legend_title,
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
theme(legend.title = element_markdown()) +
facet_grid(month ~ product) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)pco2_product_map_monthly_anomaly_ensemble <-
pco2_product_map_monthly_anomaly %>%
filter(year == 2023,
product %in% pco2_product_list) %>%
fgroup_by(name, lon, lat, month) %>%
fsummarise(
resid_sd = fsd(resid),
resid_mean = fmean(resid),
n = fnobs(resid)
) %>%
filter(n == length(pco2_product_list)) %>%
select(-n)
pco2_product_map_monthly_anomaly_ensemble_coarse <-
m_grid_horizontal_coarse(pco2_product_map_monthly_anomaly_ensemble) %>%
fgroup_by(name, month, lon_grid, lat_grid) %>%
fsummarise(resid_sd_coarse = fmean(resid_sd, na.rm = TRUE),
resid_mean_coarse = fmean(resid_mean, na.rm = TRUE)) %>%
rename(lon = lon_grid, lat = lat_grid)
pco2_product_map_monthly_anomaly_ensemble <-
left_join(
pco2_product_map_monthly_anomaly_ensemble,
pco2_product_map_monthly_anomaly_ensemble_coarse
)
pco2_product_map_monthly_anomaly_ensemble %>%
filter(name %in% name_core) %>%
mutate(month = as.character(month),
month = fct_inorder(month)) %>%
group_split(name) %>%
head(1) %>%
map(
~ p_map_mdim_robinson(
df = .x,
var = "resid_mean",
dim_wrap = "month",
legend_title = labels_breaks(.x %>% distinct(name))$i_legend_title,
breaks = labels_breaks(.x %>% distinct(name))$i_breaks
)
)[[1]]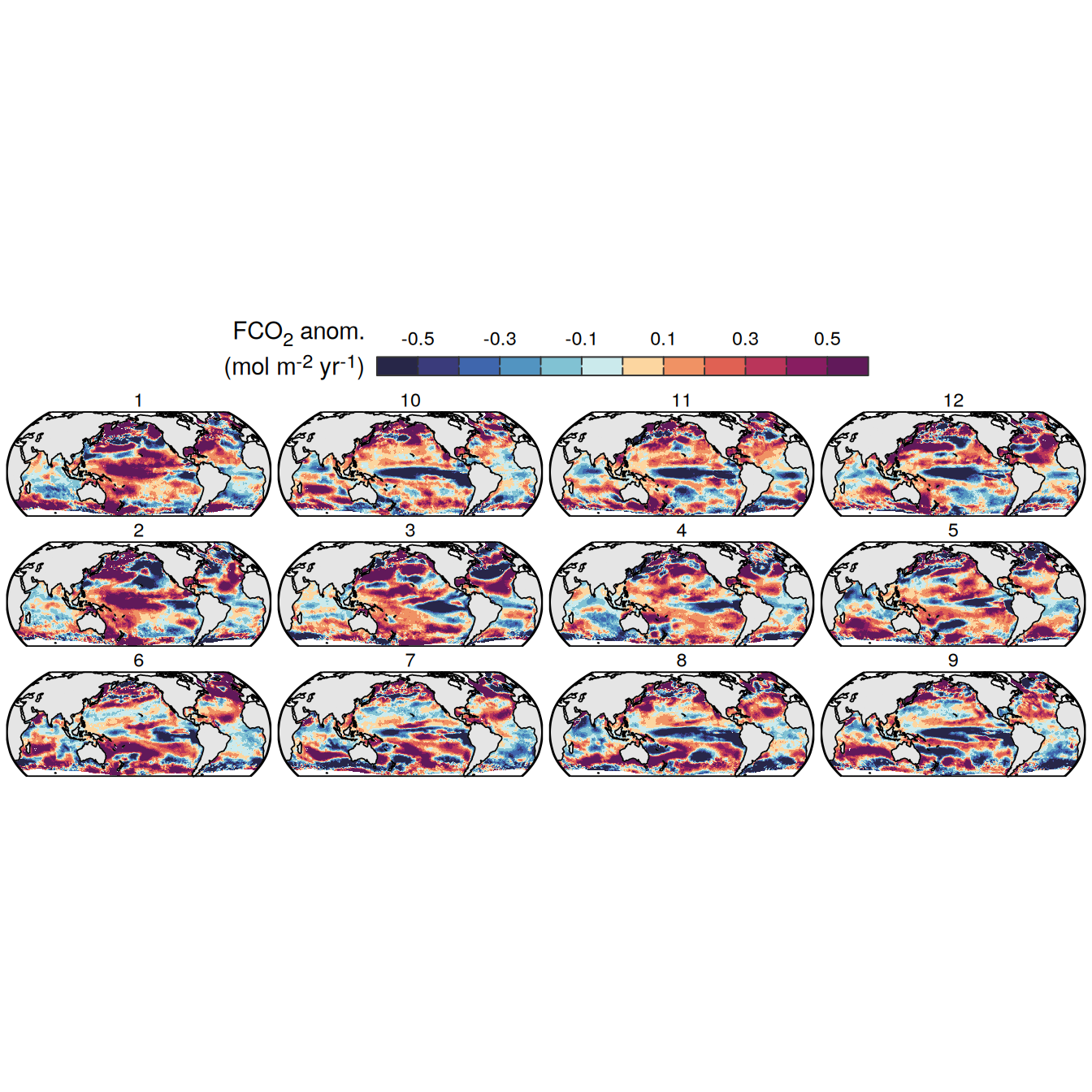
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
rm(
pco2_product_map_monthly_anomaly_ensemble,
pco2_product_map_monthly_anomaly_ensemble_coarse
)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 3261392 174.2 6540298 349.3 6540298 349.3
Vcells 275533475 2102.2 607749785 4636.8 607749785 4636.8fCO2 decomposition
pco2_product_map_monthly_fCO2_decomposition <-
inner_join(pco2_product_map_monthly_fCO2_decomposition,
biome_mask_print)pco2_product_map_monthly_fCO2_decomposition %>%
filter(year == 2023) %>%
group_split(product) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid)) +
labs(title = .x$product) +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("sfco2"),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
facet_grid(month ~ name,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)
pco2_product_map_monthly_fCO2_decomposition %>%
filter(year == 2023,
product %in% pco2_product_list) %>%
group_by(name, lon, lat, month) %>%
summarize(
resid_sd = sd(resid),
resid_mean = mean(resid),
n = n()
) %>%
ungroup() %>%
filter(n == length(pco2_product_list)) %>%
select(-n) %>%
mutate(product = "Ensemble mean") %>%
group_split(product) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid_mean)) +
# geom_point(
# data = .x %>% filter(abs(resid_mean) < resid_sd),
# aes(lon, lat, shape = "Ensemble mean\n< StDev"),
# col = "grey"
# ) +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("sfco2"),,
limits = c(quantile(.x$resid_mean, .01), quantile(.x$resid_mean, .99)),
oob = squish
) +
scale_shape_manual(values = 46, name = "") +
facet_grid(month ~ name,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)pco2_product_map_annual_fCO2_decomposition <-
pco2_product_map_monthly_fCO2_decomposition %>%
select(product, year, lat, lon, name, resid) %>%
fgroup_by(product, year, lat, lon, name) %>%
fmean()
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 3233585 172.7 6540298 349.3 6540298 349.3
Vcells 250136813 1908.4 607749785 4636.8 607749785 4636.8pco2_product_map_annual_fCO2_decomposition %>%
filter(year == 2023) %>%
select(-year) %>%
relocate(lon, lat) %>%
# mutate(name = str_remove(name, "sfco2_")) %>%
p_map_mdim_robinson(
var = "resid",
dim_col = "name",
dim_row = "product",
legend_title = labels_breaks("sfco2")$i_legend_title,
breaks = 2 * (labels_breaks("sfco2")$i_breaks),
n_labels = 2
)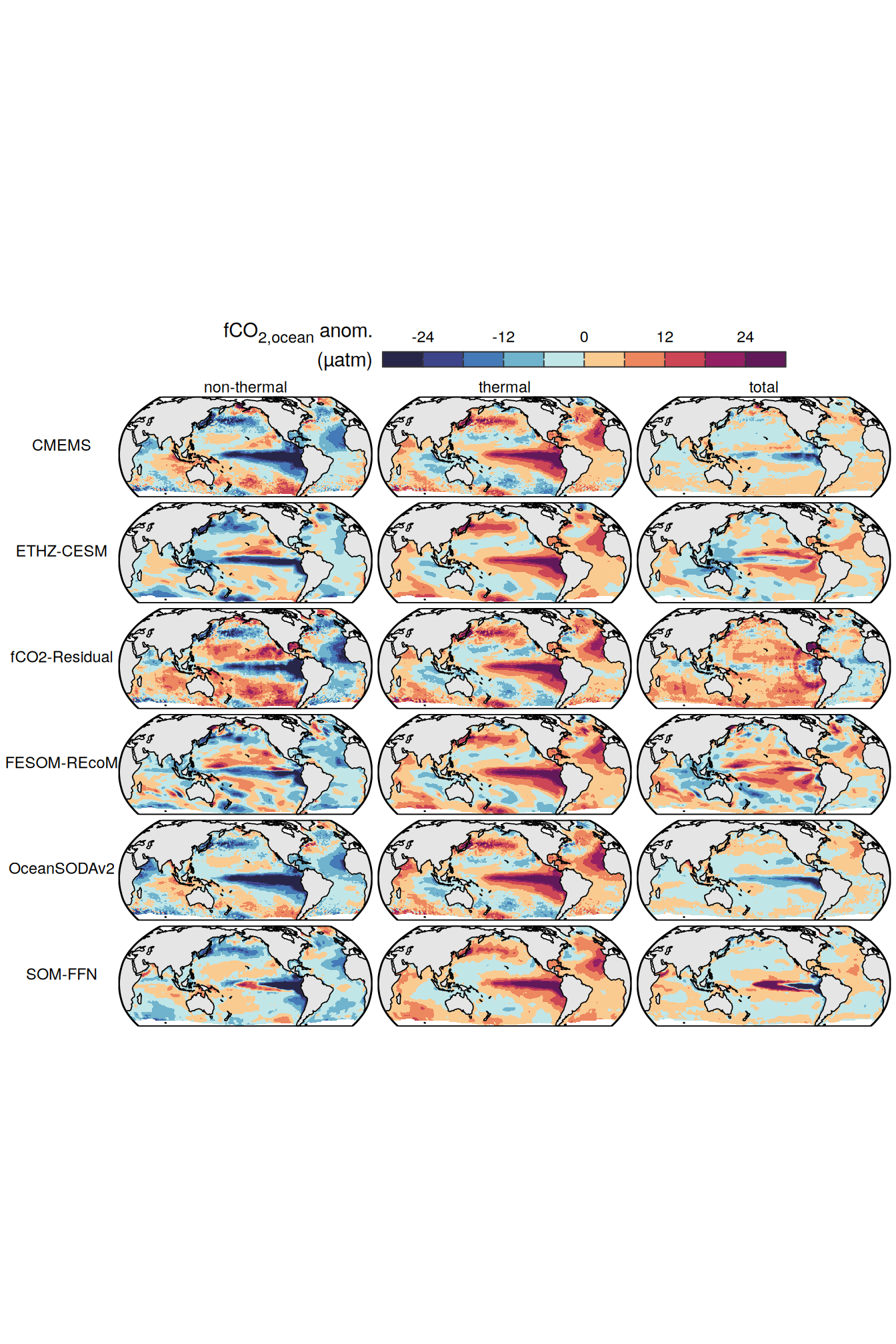
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| 681629f | jens-daniel-mueller | 2024-08-26 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
pco2_product_map_annual_fCO2_decomposition_ensemble <-
pco2_product_map_annual_fCO2_decomposition %>%
filter(product %in% pco2_product_list, year == 2023) %>%
group_by(name, lon, lat) %>%
summarize(resid_sd = sd(resid),
resid_mean = mean(resid),
n = n()) %>%
ungroup() %>%
filter(n == length(pco2_product_list)) %>%
select(-n)
pco2_product_map_annual_fCO2_decomposition_ensemble_coarse <-
m_grid_horizontal_coarse(pco2_product_map_annual_fCO2_decomposition_ensemble) %>%
fgroup_by(name, lon_grid, lat_grid) %>%
fsummarise(resid_sd_coarse = fmean(resid_sd, na.rm = TRUE),
resid_mean_coarse = fmean(resid_mean, na.rm = TRUE)) %>%
rename(lon = lon_grid, lat = lat_grid)
pco2_product_map_annual_fCO2_decomposition_ensemble_uncertainty <-
pco2_product_map_annual_fCO2_decomposition_ensemble_coarse %>%
mutate(signif_single = if_else(abs(resid_mean_coarse) < resid_sd_coarse, 0, 1)) %>%
select(lon, lat, name, signif_single) %>%
st_as_sf(coords = c("lon", "lat"), crs = "+proj=longlat")
pco2_product_map_annual_fCO2_decomposition_ensemble %>%
select(lon, lat, name, resid_mean) %>%
mutate(name = fct_relevel(name,
c("sfco2_therm", "sfco2_nontherm"))) %>%
p_map_mdim_robinson(
df_uncertainty = pco2_product_map_annual_fCO2_decomposition_ensemble_uncertainty,
var = "resid_mean",
legend_title = labels_breaks("sfco2")$i_legend_title,
breaks = 2*(labels_breaks("sfco2")$i_breaks),
dim_wrap = "name",
n_col = 1,
n_labels = 2
)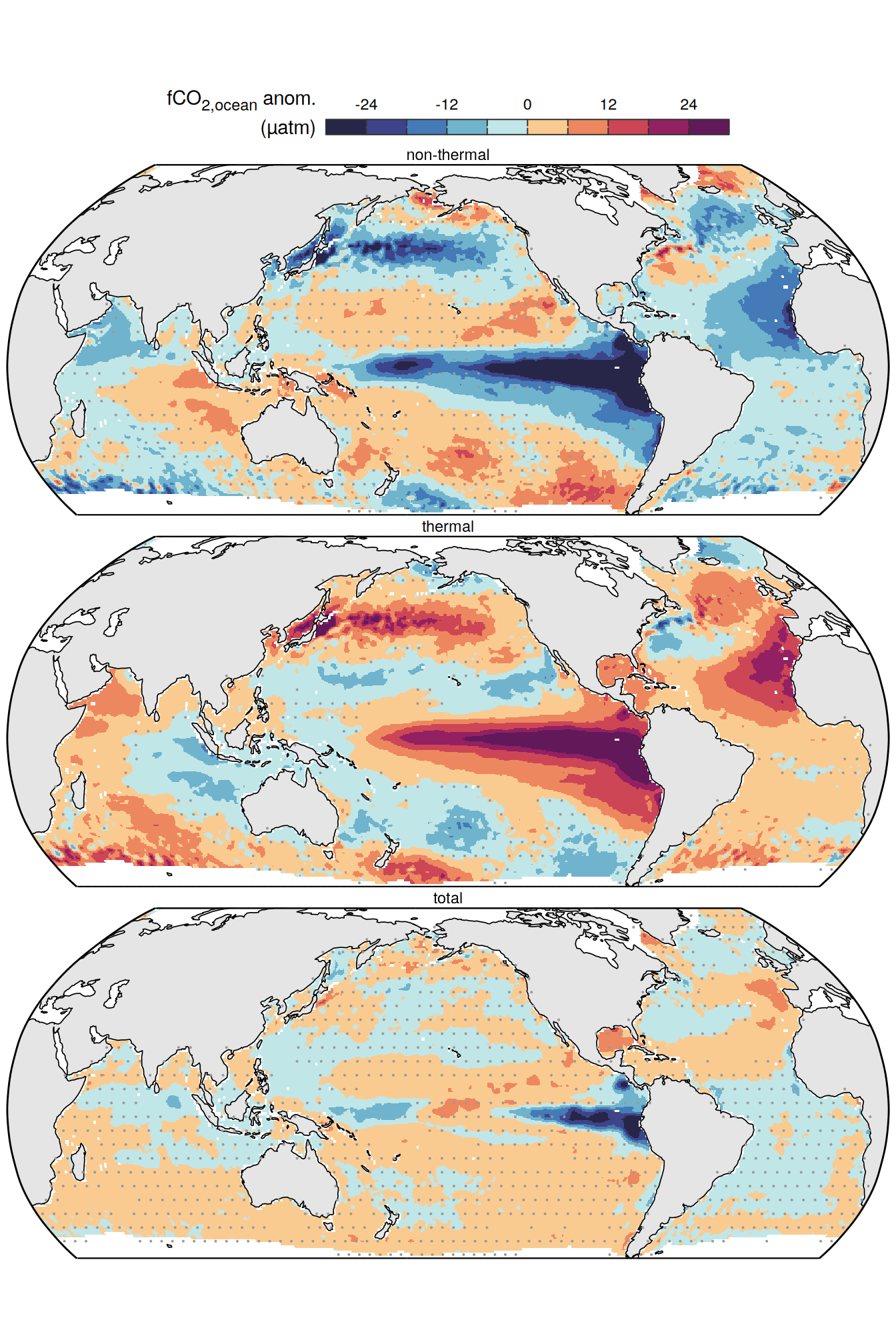
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 1eefab2 | jens-daniel-mueller | 2024-05-21 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
ggsave(width = 5,
height = 7,
dpi = 600,
filename = "../output/map_anomaly_fco2_decomposition_ensemble_mean_pco2_products.jpg")Flux attribution
pco2_product_map_monthly_flux_attribution <-
inner_join(pco2_product_map_monthly_flux_attribution, biome_mask_print)# pco2_product_map_monthly_flux_attribution <-
# flux_attribution(pco2_product_map_monthly_anomaly,
# year, month, lon, lat)
pco2_product_map_monthly_flux_attribution %>%
filter(year == 2023) %>%
drop_na() %>%
group_split(product) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid)) +
labs(subtitle = .x$product) +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("fgco2"),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
theme(legend.title = element_markdown(),
legend.position = "bottom") +
facet_grid(month ~ name,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top",
strip.text.x.top = element_markdown())
)
pco2_product_map_monthly_flux_attribution %>%
filter(year == 2023) %>%
drop_na() %>%
filter(product %in% pco2_product_list) %>%
group_by(name, lon, lat, month) %>%
summarize(
resid_sd = sd(resid),
resid_mean = mean(resid),
n = n()
) %>%
ungroup() %>%
filter(n == length(pco2_product_list)) %>%
select(-n) %>%
mutate(product = "Ensemble mean") %>%
drop_na() %>%
group_split(product) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid_mean)) +
# geom_point(data = .x %>% filter(abs(resid_mean) < resid_sd),
# aes(lon, lat, shape = "Ensemble mean\n< StDev"))+
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("fgco2"),
limits = c(quantile(.x$resid_mean, .01), quantile(.x$resid_mean, .99)),
oob = squish
)+
scale_shape_manual(values = 46, name = "") +
theme(legend.title = element_markdown(),
legend.position = "bottom") +
facet_grid(month ~ name,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.title = element_markdown(),
legend.position = "top",
strip.text.x.top = element_markdown()
)
)pco2_product_map_annual_flux_attribution <-
pco2_product_map_monthly_flux_attribution %>%
group_by(product, year, lat, lon, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup()
pco2_product_map_annual_flux_attribution %>%
filter(year == 2023) %>%
select(-year) %>%
relocate(lon, lat) %>%
# mutate(name = str_remove_all(name, "_")) %>%
p_map_mdim_robinson(
var = "resid",
dim_row = "product",
dim_col = "name",
legend_title = labels_breaks("fgco2")$i_legend_title,
breaks = labels_breaks("fgco2")$i_breaks,
n_labels = 2
)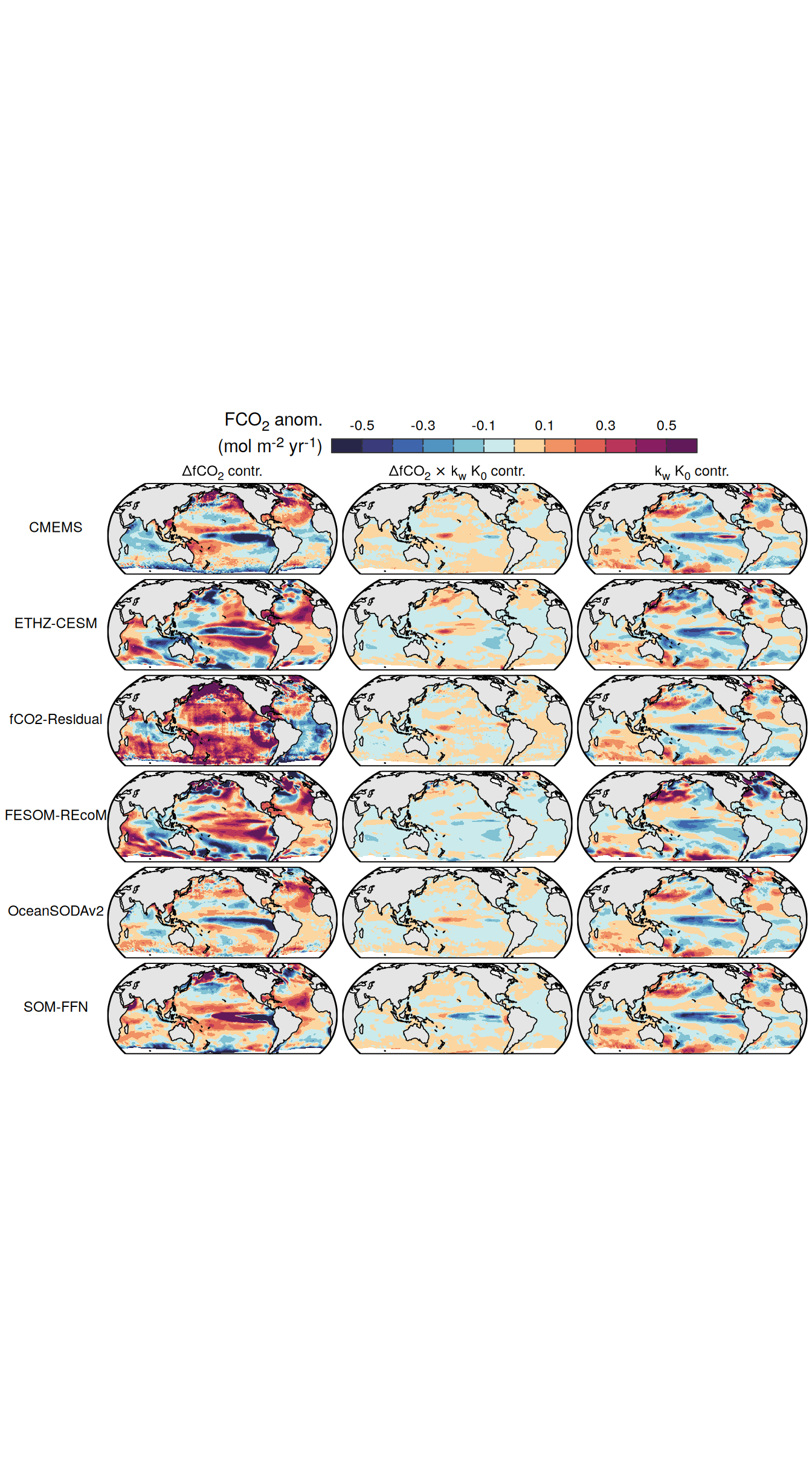
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 681629f | jens-daniel-mueller | 2024-08-26 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
pco2_product_map_annual_flux_attribution_ensemble <-
pco2_product_map_annual_flux_attribution %>%
filter(year == 2023,
product %in% pco2_product_list) %>%
group_by(name, lon, lat) %>%
summarize(
resid_sd = sd(resid),
resid_mean = mean(resid),
n = n()
) %>%
ungroup() %>%
filter(n == length(pco2_product_list)) %>%
select(-n) %>%
drop_na()
pco2_product_map_annual_flux_attribution_ensemble_coarse <-
m_grid_horizontal_coarse(pco2_product_map_annual_flux_attribution_ensemble) %>%
fgroup_by(name, lon_grid, lat_grid) %>%
fsummarise(resid_sd_coarse = fmean(resid_sd, na.rm = TRUE),
resid_mean_coarse = fmean(resid_mean, na.rm = TRUE)) %>%
rename(lon = lon_grid, lat = lat_grid)
pco2_product_map_annual_flux_attribution_ensemble_uncertainty <-
pco2_product_map_annual_flux_attribution_ensemble_coarse %>%
mutate(signif_single = if_else(abs(resid_mean_coarse) < resid_sd_coarse, 0, 1)) %>%
select(lon, lat, name, signif_single) %>%
st_as_sf(coords = c("lon", "lat"), crs = "+proj=longlat")
pco2_product_map_annual_flux_attribution_ensemble %>%
select(lon, lat, name, resid_mean) %>%
p_map_mdim_robinson(
df_uncertainty = pco2_product_map_annual_flux_attribution_ensemble_uncertainty,
var = "resid_mean",
legend_title = labels_breaks("fgco2")$i_legend_title,
breaks = labels_breaks("fgco2")$i_breaks,
dim_wrap = "name",
n_col = 1,
n_labels = 2
)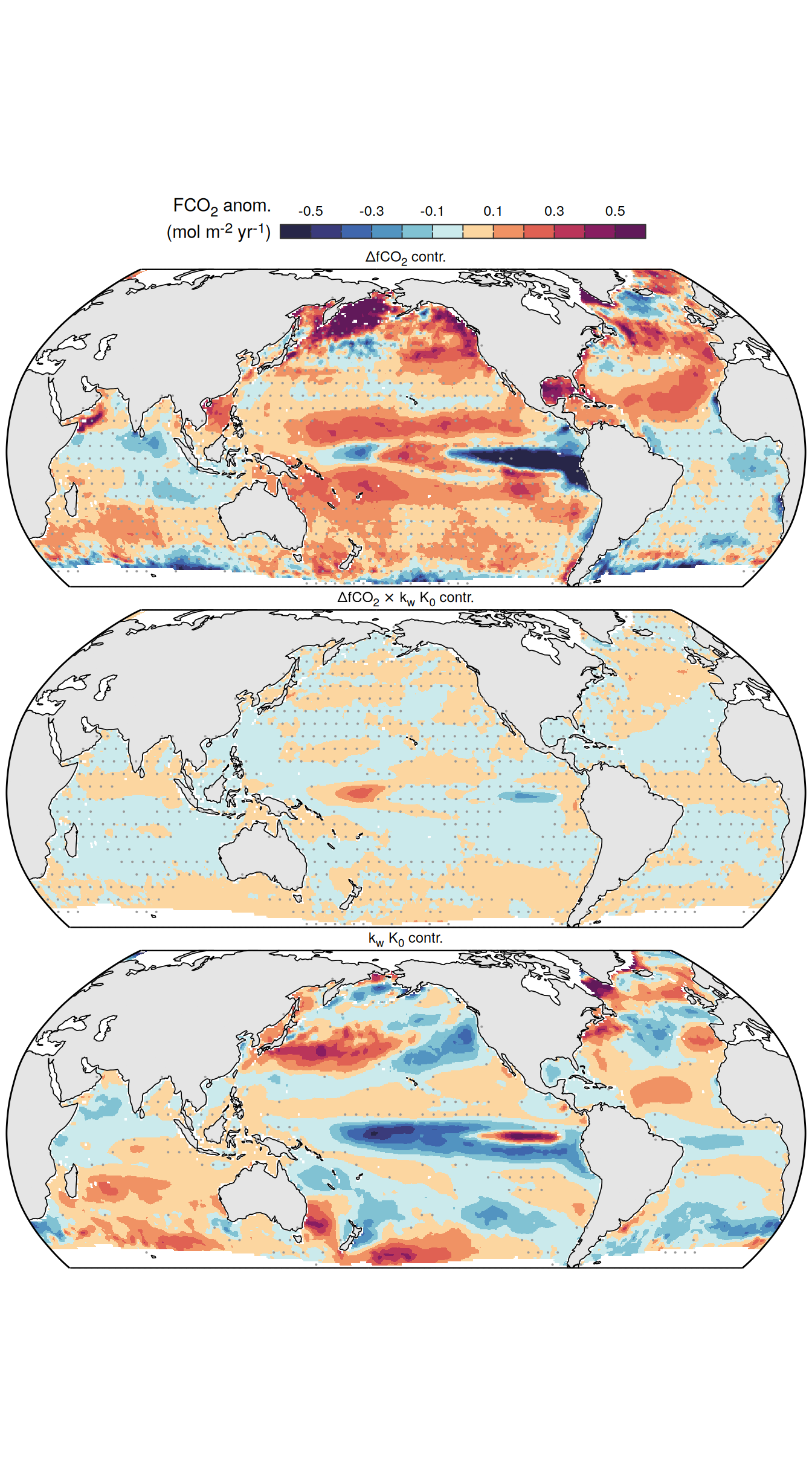
| Version | Author | Date |
|---|---|---|
| 17f843d | jens-daniel-mueller | 2025-03-10 |
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 1eefab2 | jens-daniel-mueller | 2024-05-21 |
| a29d870 | jens-daniel-mueller | 2024-05-16 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
ggsave(width = 5,
height = 7,
dpi = 600,
filename = "../output/map_anomaly_flux_attribution_ensemble_mean_pco2_products.jpg")Hovmoeller plots
The following Hovmoeller plots show the anomalies from the prediction of a linear/quadratic fit to the data from 1990 to 2022.
Hovmoeller plots are presented as monthly means. Note that the predictions for the monthly Hovmoeller plots are done individually for each month, such the mean seasonal anomaly from the annual mean is removed.
Monthly means
Anomalies
pco2_product_hovmoeller_monthly_anomaly %>%
filter(name %in% name_core) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(decimal, lat, fill = resid)) +
geom_raster() +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name))$i_legend_title,
limits = c(quantile(.x$resid,.01),quantile(.x$resid,.99)),
oob = squish
) +
theme(legend.title = element_markdown()) +
coord_cartesian(expand = 0) +
labs(title = "Monthly mean anomalies",
y = "Latitude") +
theme(axis.title.x = element_blank()) +
facet_wrap(~ product, ncol = 1)
)pco2_product_hovmoeller_monthly_anomaly_ensemble <-
pco2_product_hovmoeller_monthly_anomaly %>%
group_by(name, decimal, lat) %>%
summarize(
resid_range = max(resid) - min(resid),
resid_mean = mean(resid),
n = n()
) %>%
ungroup() %>%
filter(n > 1)
pco2_product_hovmoeller_monthly_anomaly_ensemble %>%
mutate(product = "Ensemble mean") %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(decimal, lat, fill = resid_mean)) +
geom_raster() +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name))$i_legend_title,
limits = c(quantile(.x$resid_mean, .01), quantile(.x$resid_mean, .99)),
oob = squish
) +
theme(legend.title = element_markdown()) +
coord_cartesian(expand = 0) +
labs(title = "Monthly mean anomalies",
y = "Latitude") +
theme(axis.title.x = element_blank()) +
facet_wrap( ~ product, ncol = 1)
)left_join(
pco2_product_hovmoeller_monthly_anomaly_ensemble,
pco2_product_hovmoeller_monthly_anomaly
) %>%
mutate(resid_offset = resid - resid_mean) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(decimal, lat, fill = resid_offset)) +
geom_raster() +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name))$i_legend_title,
limits = c(quantile(.x$resid_mean, .01), quantile(.x$resid_mean, .99)),
oob = squish
) +
theme(legend.title = element_markdown()) +
coord_cartesian(expand = 0)+
labs(title = "Monthly offset from ensemble mean anomalies",
y = "Latitude") +
theme(axis.title.x = element_blank()) +
facet_wrap( ~ product, ncol = 1)
)Regional means and integrals
The following plots show biome-, super biome- or global- averaged/integrated values of each variable as provided through the fCO2 product, represented here as the anomalies from the prediction of a linear/quadratic fit to the data from 1990 to 2022.
Anomalies are presented relative to the predicted annual mean of each year, hence preserving the seasonality.
Annual anomalies
pco2_product_biome_annual_anomaly_ensemble <-
pco2_product_biome_annual_anomaly %>%
filter(product %in% pco2_product_list) %>%
group_by(year, name, biome) %>%
summarise(resid_sd = sd(resid),
resid = mean(resid),
value = mean(value),
fit = mean(fit)) %>%
ungroup()
# fit lin. reg. model fgco2 anomaly vs SST anomaly per biome
# excluding anomaly year from fit
lm_fgco2_sst <- pco2_product_biome_annual_anomaly %>%
filter(
name %in% c("fgco2_int", "temperature"),
biome == "Global non-polar",
product %in% pco2_product_list
) %>%
select(year, product, name, resid) %>%
pivot_wider(values_from = resid) %>%
nest(data = -product) %>%
mutate(fit = map(data, ~ lm(
formula = fgco2_int ~ temperature,
data = .x %>% filter(year != 2023)
)))
# predict fgco2 anomaly from SST anomaly per biome
# include anomaly year for prediction
lm_fgco2_sst <- lm_fgco2_sst %>%
mutate(resid = map2(fit, data, predict)) %>%
unnest(c(data, resid))
# extract regression coefficients
lm_fgco2_sst <- lm_fgco2_sst %>%
mutate(coeff = map(fit, coefficients)) %>%
select(-fit) %>%
rename(temperature_obs = temperature) %>%
unnest_wider(coeff) %>%
rename(intercept = `(Intercept)`,
slope = temperature,
temperature = temperature_obs)
lm_fgco2_sst %>%
filter(year == 2023) %>%
select(-fgco2_int) %>%
mutate(across(c(slope, temperature, resid), ~ round(.x, 2)),
across(c(intercept), ~ signif(.x, 2))) %>%
write_csv("../output/lm_fgco2_sst.csv")
# compute unexpected (predicted vs observed) flux anomalies
lm_fgco2_sst %>%
ggplot(aes(year, fgco2_int-resid, col = product)) +
geom_hline(yintercept = 0) +
geom_path() +
labs(title = "Unexpected residuals in ocean carbon sink",
subtitle = "Linear regression baseline + global mean SST anomaly")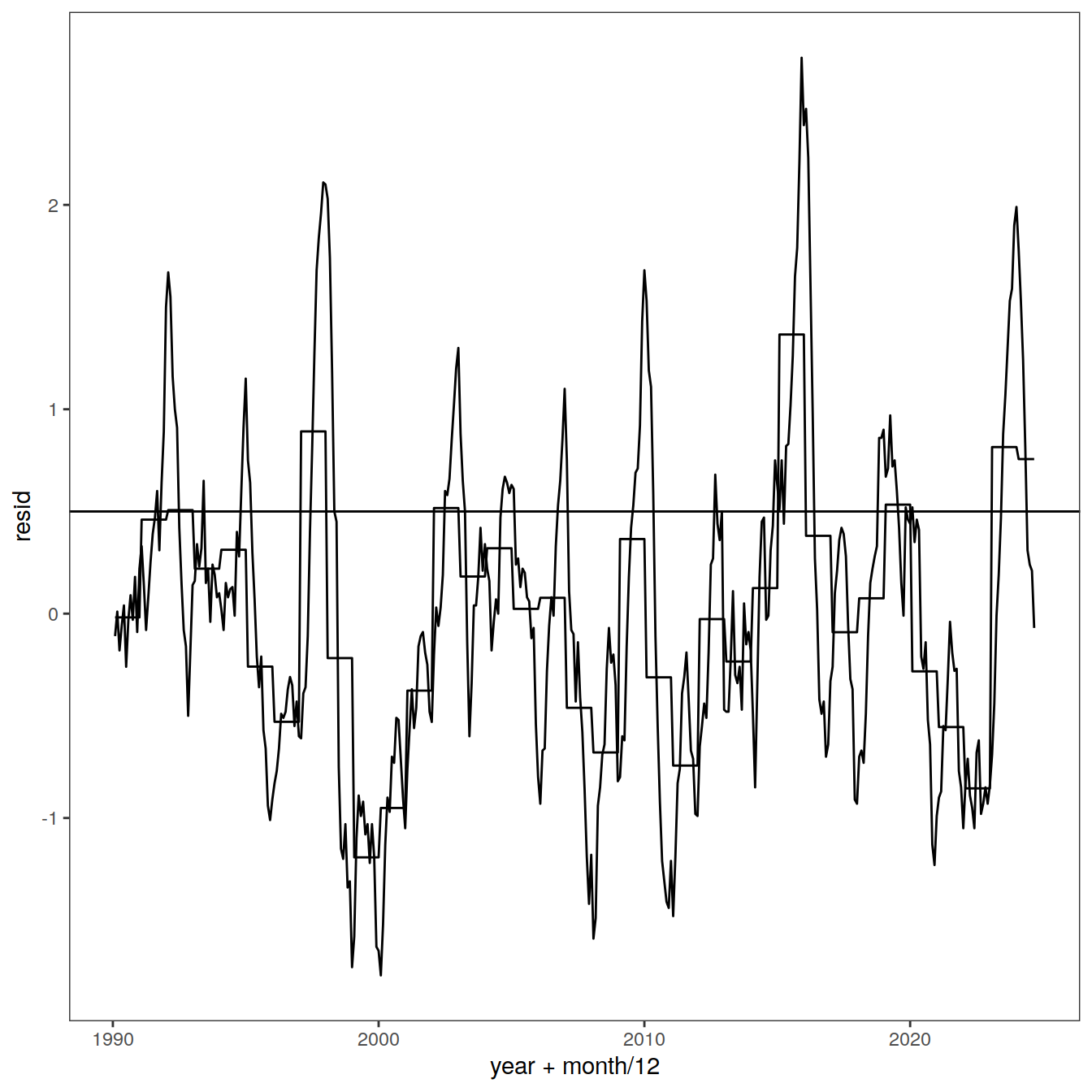
| Version | Author | Date |
|---|---|---|
| b031240 | jens-daniel-mueller | 2025-03-11 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 12f6571 | jens-daniel-mueller | 2024-05-22 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| 1eefab2 | jens-daniel-mueller | 2024-05-21 |
pco2_product_biome_annual_anomaly_lm_fgco2_sst <-
inner_join(
pco2_product_biome_annual_anomaly %>%
filter(
name %in% c("fgco2_int"),
biome == "Global non-polar",
product %in% pco2_product_list
) %>%
select(year, product, baseline = fit),
lm_fgco2_sst %>%
select(year, product, resid)
)
lm_fgco2_sst <-
lm_fgco2_sst %>%
group_by(year) %>%
summarise(
resid_sd = sd(resid),
resid_mean = mean(resid),
unexplained_sd = sd(fgco2_int - resid),
unexplained_mean = mean(fgco2_int - resid),
temperature_sd = sd(temperature),
temperature_mean = mean(temperature)
) %>%
ungroup()
lm_fgco2_sst %>%
ggplot(aes(year, unexplained_mean,
ymin = unexplained_mean - unexplained_sd,
ymax = unexplained_mean + unexplained_sd)) +
geom_hline(yintercept = 0) +
geom_ribbon(alpha = 0.2) +
geom_path() +
labs(title = "Unexpected residuals in ocean carbon sink",
subtitle = "Linear regression baseline + global mean SST anomaly")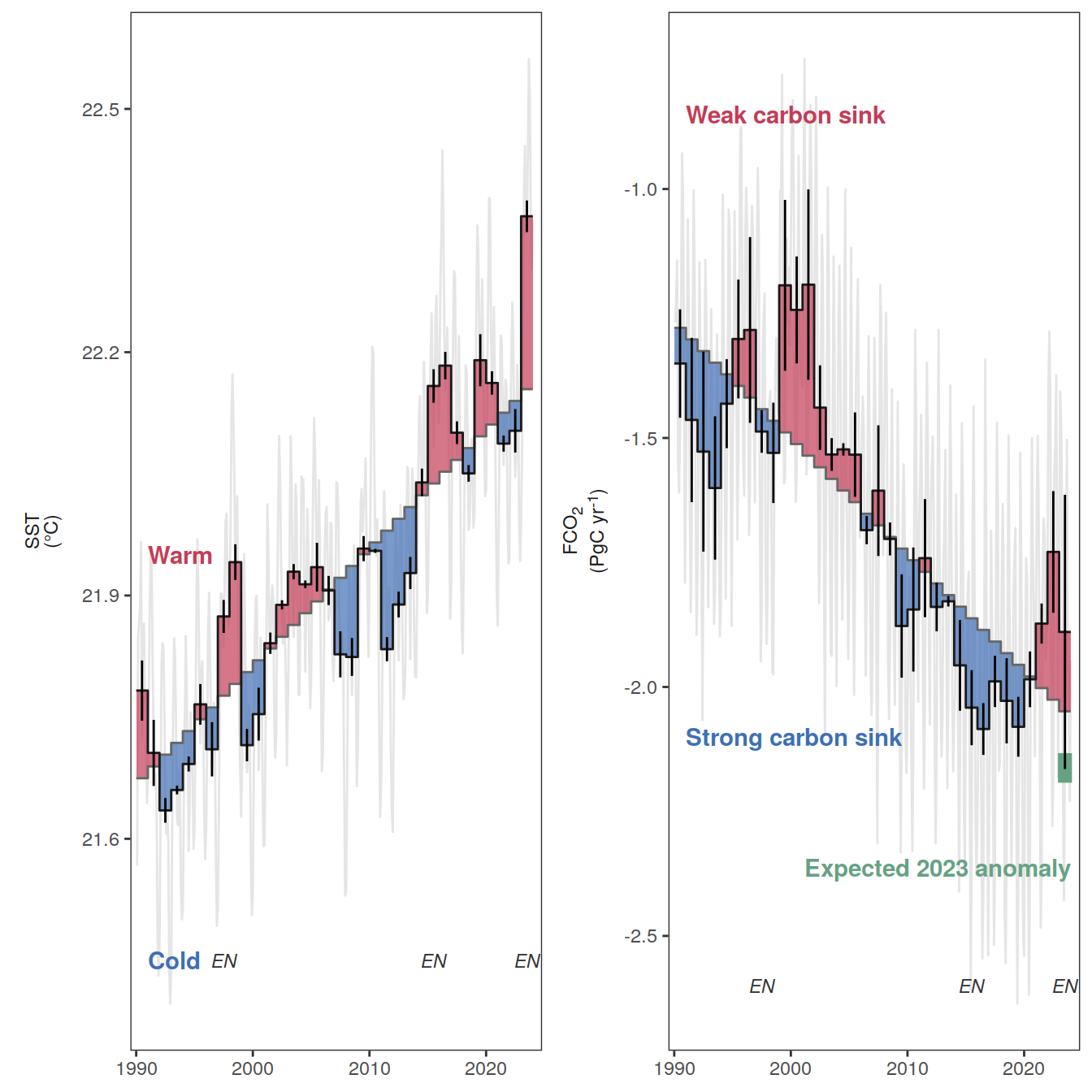
| Version | Author | Date |
|---|---|---|
| b031240 | jens-daniel-mueller | 2025-03-11 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 713d232 | jens-daniel-mueller | 2024-09-12 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| ac8cc7e | jens-daniel-mueller | 2024-05-24 |
pco2_product_biome_annual_anomaly_ensemble_lm_fgco2_sst <-
inner_join(
lm_fgco2_sst %>%
select(-starts_with("unex")),
pco2_product_biome_annual_anomaly_ensemble %>%
filter(name %in% c("fgco2_int"), biome == "Global non-polar") %>%
select(year, name, fit)
) %>%
mutate(fgco2_predict = resid_mean + fit) %>%
select(-fit)
nino_sst %>%
filter(year >= 1990) %>%
ggplot(aes(year + month/12, resid)) +
geom_hline(yintercept = 0.5) +
geom_path(aes(col = "Monthly mean")) +
geom_path(data = . %>%
group_by(year) %>%
mutate(resid = mean(resid)) %>%
ungroup(),
aes(col = "Annual mean")) +
labs(y = "Nino 3.4 SST anomaly (°C)") +
theme(axis.title.x = element_blank())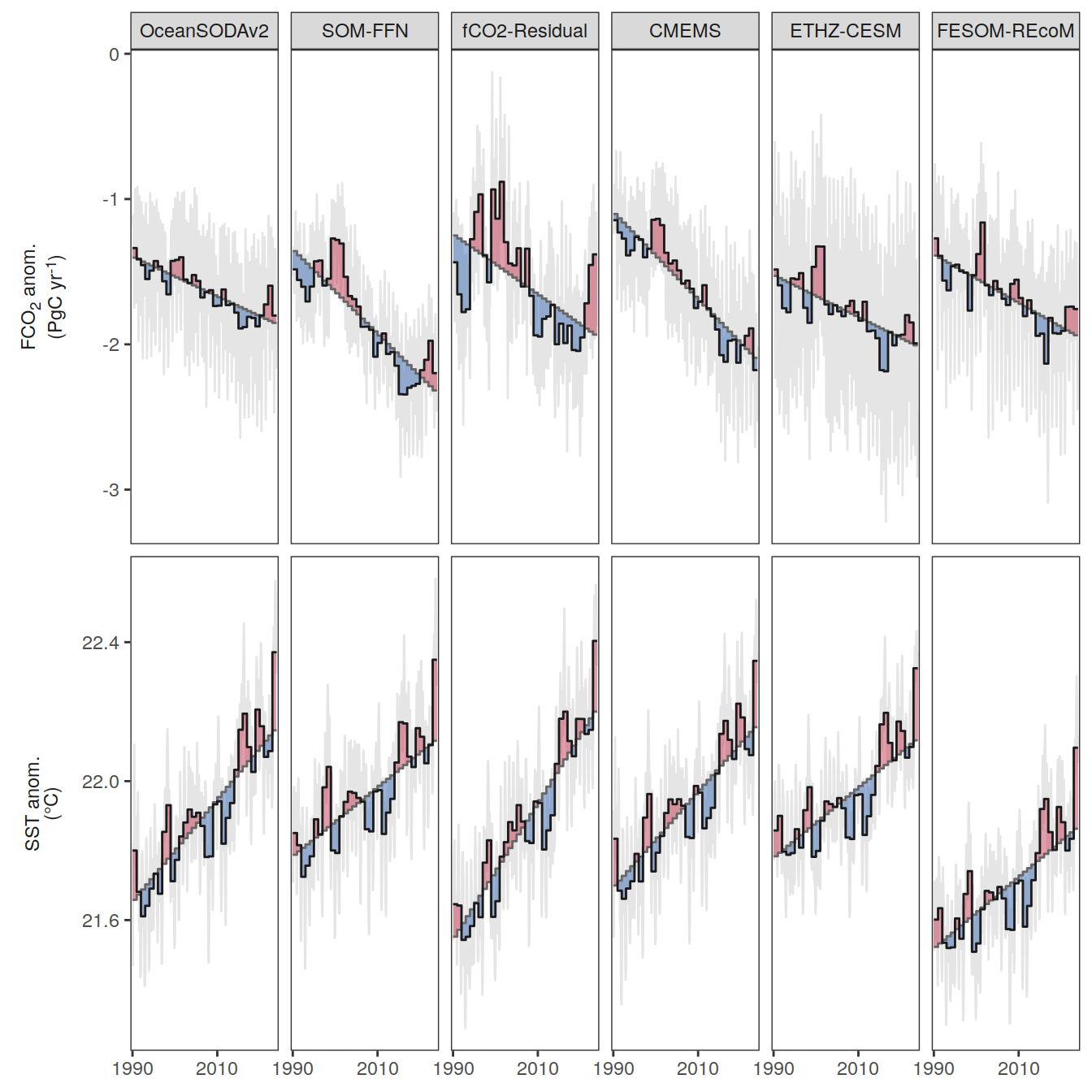
| Version | Author | Date |
|---|---|---|
| b031240 | jens-daniel-mueller | 2025-03-11 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| ac8cc7e | jens-daniel-mueller | 2024-05-24 |
bind_rows(
pco2_product_biome_annual_anomaly_ensemble,
pco2_product_biome_annual_anomaly_ensemble %>%
filter(year == max(year)) %>%
mutate(year = year + 1) %>%
select(-c(resid, resid_sd))
) %>%
filter(name %in% c("fgco2_int", "temperature"), biome == "Global non-polar") %>%
mutate(name = fct_rev(as.factor(name))) %>%
ggplot() +
geom_path(
data = pco2_product_biome_monthly_anomaly %>%
filter(
product %in% pco2_product_list,
name %in% c("fgco2_int", "temperature"),
biome == "Global non-polar"
) %>%
group_by(year, month, name, biome) %>%
summarise(value = mean(value)) %>%
ungroup(),
aes(year + month / 12, value),
col = "grey90"
) +
geom_rect(
data = pco2_product_biome_annual_anomaly_ensemble_lm_fgco2_sst %>%
filter(year %in% 2023),
aes(xmin = year, xmax = year + 1, ymin = fgco2_predict - resid_sd,
ymax = fgco2_predict + resid_sd),
fill = trend_color, col = trend_color
) +
geom_text(
data = pco2_product_biome_annual_anomaly_ensemble_lm_fgco2_sst %>%
filter(year %in% c(2023)),
aes(x = year + 1, y = fgco2_predict - 0.2, label = "Expected 2023 anomaly"),
hjust = 1,
fontface = "bold",
col = trend_color
) +
geom_text(
data = . %>%
filter(year == 1991, name == "temperature"),
aes(x = year, y = 21.95, label = "Warm"),
hjust = 0,
fontface = "bold",
col = warm_color
) +
geom_text(
data = . %>%
filter(year == 1991, name == "temperature"),
aes(x = year, y = 21.45, label = "Cold"),
hjust = 0,
fontface = "bold",
col = cold_color
) +
geom_text(
data = . %>%
filter(year == 1991, name == "fgco2_int"),
aes(x = year, y = -0.85, label = "Weak carbon sink"),
hjust = 0,
fontface = "bold",
col = warm_color
) +
geom_text(
data = . %>%
filter(year == 1991, name == "fgco2_int"),
aes(x = year, y = -2.1, label = "Strong carbon sink"),
hjust = 0,
fontface = "bold",
col = cold_color
) +
geom_text(
data = . %>%
filter(year %in% c(1997, 2015, 2023), name == "fgco2_int"),
aes(
x = year + 0.5,
y = -2.6,
label = "EN"
), size = 3, fontface = "italic", col = "grey20") +
geom_text(
data = . %>%
filter(year %in% c(1997, 2015, 2023), name == "temperature"),
aes(
x = year + 0.5,
y = 21.45,
label = "EN"
), size = 3, fontface = "italic", col = "grey20") +
geom_rect(
data = . %>% filter(year != max(year)),
aes(
xmin = year,
xmax = year + 1,
ymin = fit,
ymax = value,
fill = as.factor(sign(-resid))
),
alpha = 0.7
) +
geom_step(aes(year, fit, col = "Baseline")) +
geom_step(aes(year, value, col = "Observed")) +
geom_linerange(aes(
x = year + 0.5,
ymin = value - resid_sd,
ymax = value + resid_sd,
linetype = "Product SD"
)) +
scale_color_manual(values = c("grey40", "grey10"), name = "Annual means") +
scale_fill_manual(
values = c(warm_color, cold_color),
labels = c("positive", "negative"),
name = "Anomalies"
) +
scale_linetype(name = "Anomaly uncertainty") +
guides(
color = guide_legend(order = 1),
fill = guide_legend(order = 2),
linetype = guide_legend(order = 3)
) +
scale_x_continuous(limits = c(1989.5, 2024.8), expand = c(0, 0)) +
facet_wrap(
. ~ name,
scales = "free_y",
strip.position = "left",
labeller = labeller(name = x_axis_labels_abs)
# switch = "y"
)+
labs(x = "Year") +
theme(
axis.title.y = element_blank(),
axis.title.x = element_blank(),
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.position = "none",
legend.direction = "vertical"
)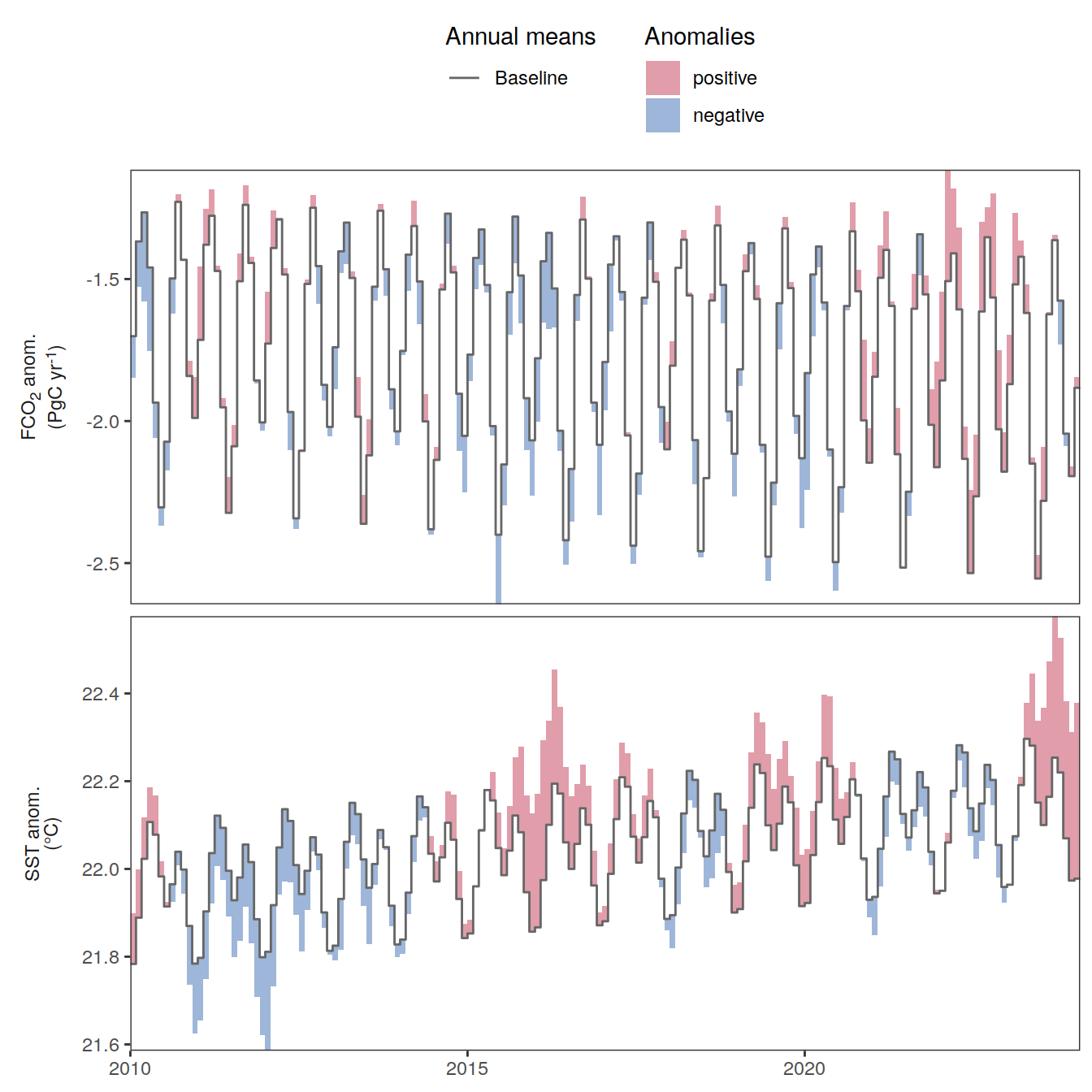
| Version | Author | Date |
|---|---|---|
| b031240 | jens-daniel-mueller | 2025-03-11 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| bf01e6c | jens-daniel-mueller | 2024-05-31 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| ac8cc7e | jens-daniel-mueller | 2024-05-24 |
ggsave(width = 10,
height = 2,
dpi = 600,
filename = "../output/timeseries_ensemble_mean_pco2_products.jpg")
bind_rows(
pco2_product_biome_annual_anomaly,
pco2_product_biome_annual_anomaly %>%
filter(year == max(year)) %>%
mutate(year = year + 1) %>%
select(-c(resid))
) %>%
filter(name %in% c("fgco2_int", "temperature"),
biome == "Global non-polar") %>%
ggplot() +
geom_path(
data = pco2_product_biome_monthly_anomaly %>%
filter(name %in% c("fgco2_int", "temperature"),
biome == "Global non-polar"),
aes(year + month / 12, value),
col = "grey90"
)+
geom_rect(
data = . %>% filter(year != max(year)),
aes(
xmin = year,
xmax = year + 1,
ymin = fit,
ymax = value,
fill = as.factor(sign(-resid))
),
alpha = 0.5
) +
geom_step(aes(year, fit, col = "Baseline")) +
geom_step(aes(year, value, col = "Observed")) +
scale_color_manual(values = c("grey40", "grey10"),
name = "Annual means") +
scale_fill_manual(
values = c(warm_color, cold_color),
labels = c("positive", "negative"),
name = "Anomalies"
) +
guides(
color = guide_legend(order = 1),
fill = guide_legend(order = 2)
) +
scale_x_continuous(limits = c(1989.5,2024.5), expand = c(0,0),
breaks = c(1990,2010)) +
facet_grid(
name ~ product,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
axis.title = element_blank(),
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.position = "none",
legend.direction = "vertical"
)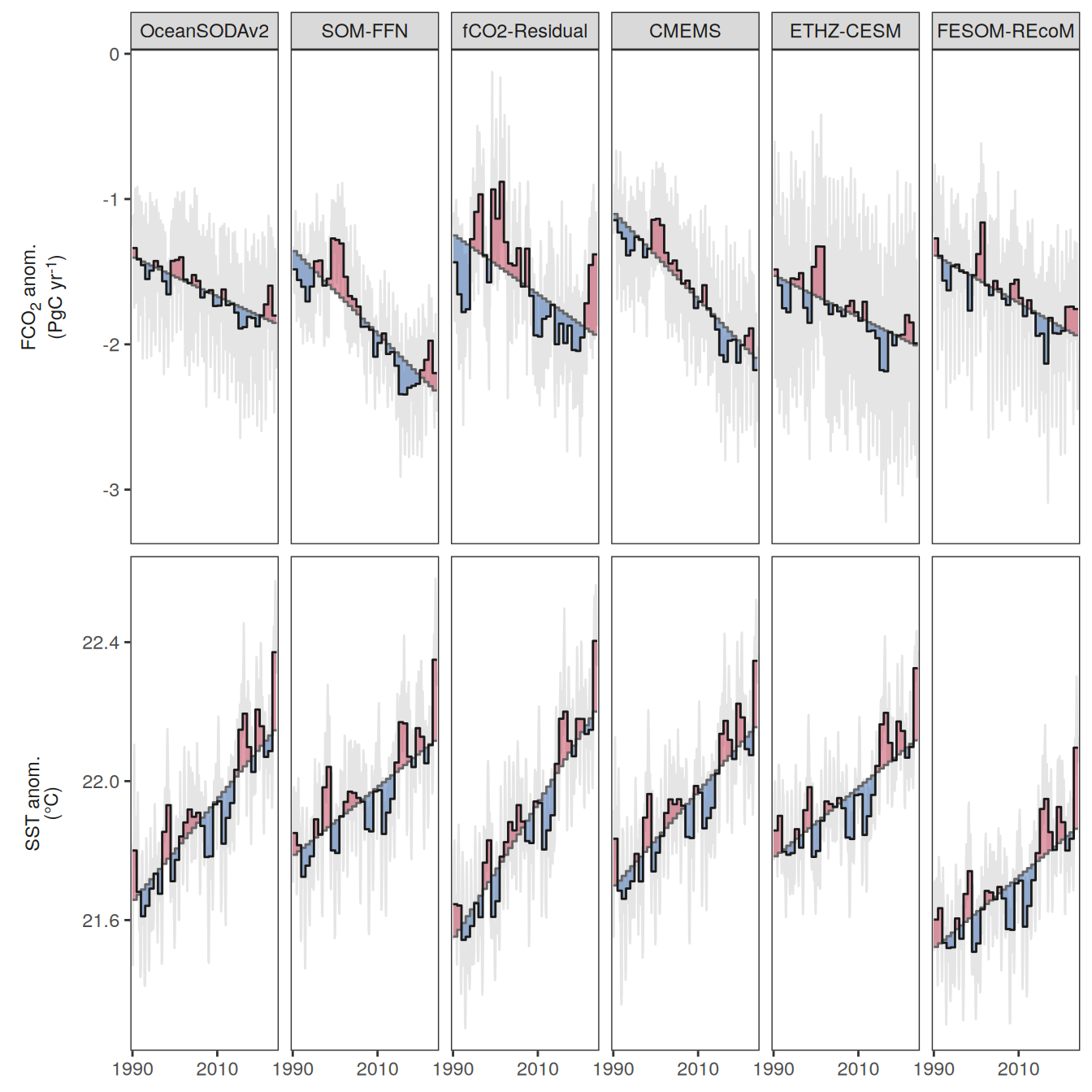
ggsave(width = 8,
height = 3,
dpi = 600,
filename = "../output/timeseries_all_products.jpg")
bind_rows(
pco2_product_biome_monthly_anomaly,
pco2_product_biome_monthly_anomaly %>%
filter(year == max(year),
month == 12) %>%
mutate(month = month + 1)
) %>%
mutate(year = year + month/12) %>%
filter(name %in% c("fgco2_int", "temperature"),
product == if(GCB_products){"OceanSODA"}else{"OceanSODAv2"},
biome == "Global non-polar",
year >= 2010) %>%
ggplot() +
geom_rect(
data = . %>% filter(year != max(year)),
aes(
xmin = year,
xmax = year + 1/12,
ymin = fit,
ymax = value,
fill = as.factor(sign(-resid))
),
alpha = 0.5
) +
geom_step(aes(year, fit, col = "Baseline")) +
scale_color_manual(values = c("grey40", "grey10"),
name = "Annual means") +
scale_fill_manual(
values = c(warm_color, cold_color),
labels = c("positive", "negative"),
name = "Anomalies"
) +
guides(color = guide_legend(order = 1),
fill = guide_legend(order = 2))+
facet_grid(
name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
coord_cartesian(expand = 0) +
theme(
axis.title = element_blank(),
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.position = "top",
legend.direction = "vertical"
)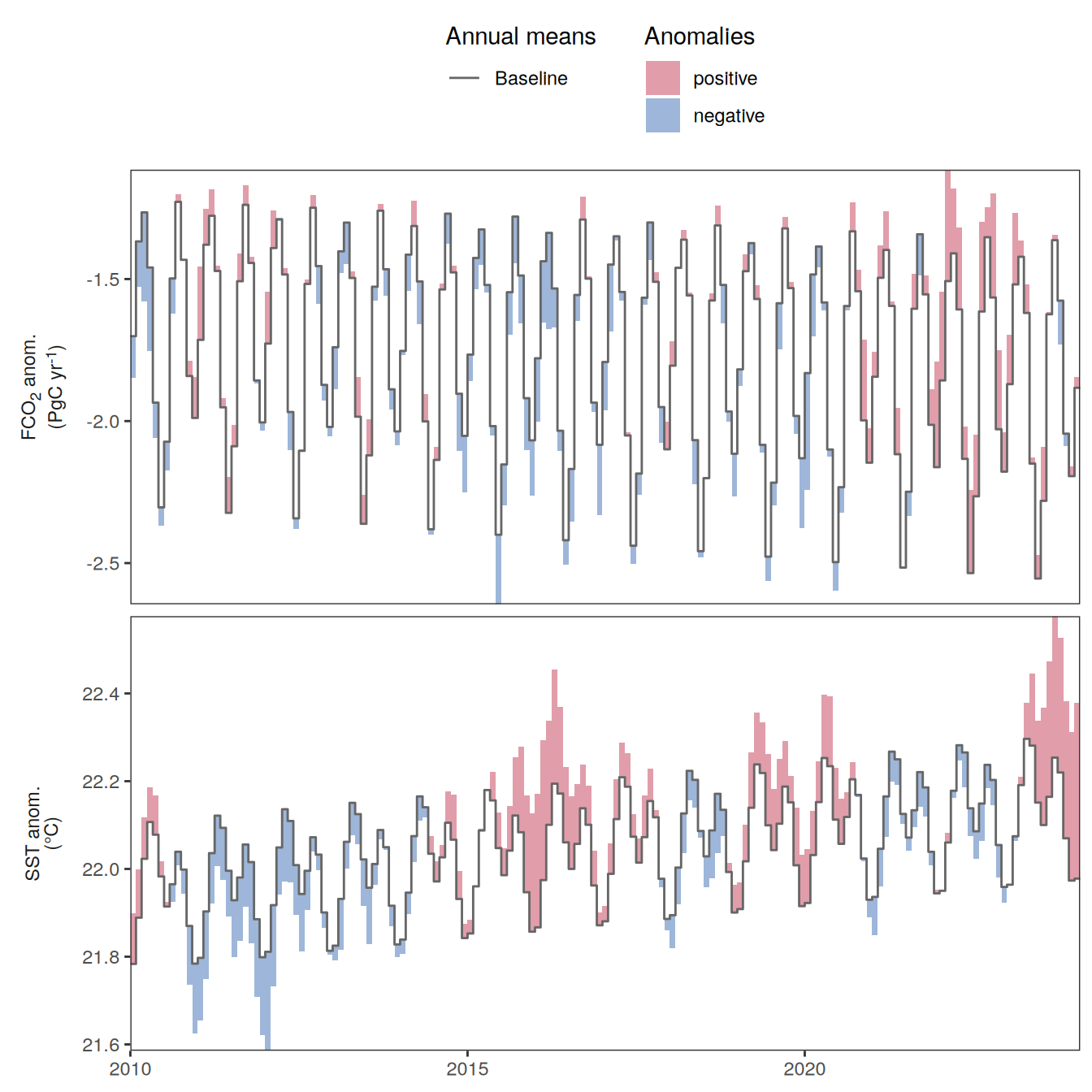
| Version | Author | Date |
|---|---|---|
| b031240 | jens-daniel-mueller | 2025-03-11 |
df_baseline <-
pco2_product_biome_annual_anomaly %>%
filter(biome == "Global non-polar",
name %in% c("fgco2_int", "temperature"),
product %in% pco2_product_list) %>%
select(product, year, name, value) %>%
pivot_wider() %>%
rename(fgco2_int_observed = fgco2_int)
df_baseline <-
inner_join(df_baseline, co2_annmean_gl %>% select(year, atmco2 = mean))
df_baseline <-
inner_join(df_baseline, co2_gr_gl %>% select(year, atmco2_gr = `ann inc`))
df_baseline <-
inner_join(df_baseline, nino_sst %>%
group_by(year) %>%
summarise(nino34 = mean(resid)) %>%
ungroup())
df_baseline <-
inner_join(
df_baseline,
pco2_product_biome_annual_anomaly_lm_fgco2_sst %>%
mutate(fgco2_int_predict_baseline_global_SST = baseline + resid) %>%
select(product, year, fgco2_int_predict_baseline_global_SST)
)
df_baseline %>%
pivot_longer(-c(year, product)) %>%
ggplot(aes(year, value)) +
geom_path() +
facet_grid(name ~ product, scales = "free_y")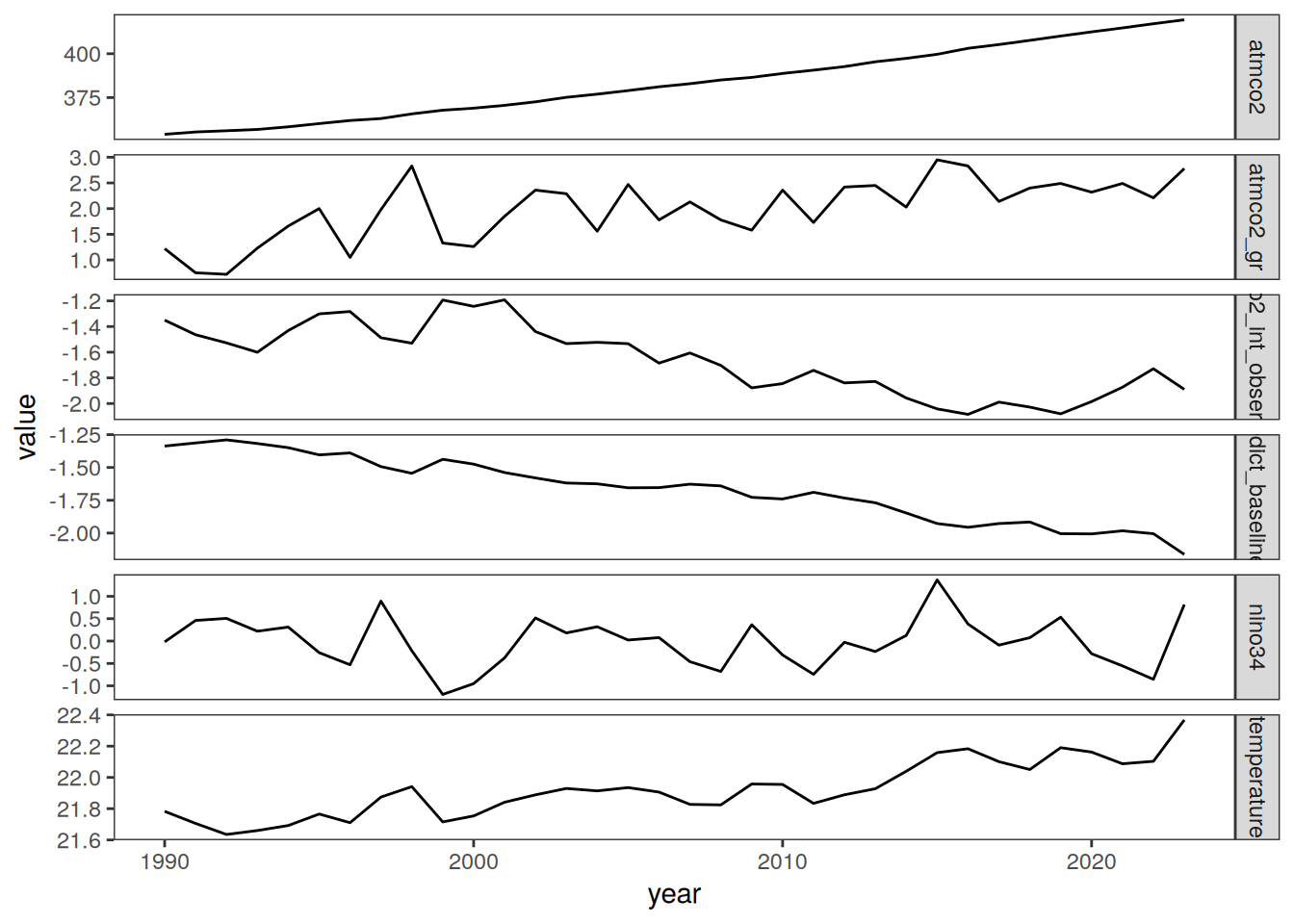
df_baseline %>%
select(-fgco2_int_predict_baseline_global_SST) %>%
pivot_longer(-c(product, year, fgco2_int_observed)) %>%
ggplot(aes(value, fgco2_int_observed)) +
geom_path(col = "grey80") +
geom_point(aes(fill = year),
shape = 21) +
scale_fill_viridis_c() +
facet_grid(product ~ name, scales = "free")
# fit lin. reg. model fgco2 anomaly vs SST anomaly per biome
# excluding anomaly year from fit
mlr <- df_baseline %>%
nest(data = -product) %>%
mutate(fit = map(data, ~ lm(
formula = fgco2_int_observed ~ atmco2 + atmco2_gr + nino34,
data = .x %>% filter(year != 2023)
)))
# predict fgco2 anomaly from SST anomaly per biome
# include anomaly year for prediction
mlr <- mlr %>%
mutate(fgco2_int_predict_atm_nino = map2(fit, data, predict)) %>%
unnest(c(data, fgco2_int_predict_atm_nino))
mlr %>%
select(product, year, starts_with("fgco2_int")) %>%
pivot_longer(starts_with("fgco2_int"),
names_prefix = "fgco2_int_") %>%
ggplot(aes(year, value, col = name)) +
geom_path() +
geom_smooth(data = . %>% filter(name == "observed",
year != 2023),
method = "lm",
se = FALSE,
fullrange = TRUE) +
scale_color_okabeito() +
facet_wrap(~ product) +
labs(y = str_remove(labels_breaks("fgco2_int")$i_legend_title, "anom.")) +
theme(axis.title.x = element_blank(),
axis.title.y = element_markdown(),
legend.title = element_blank())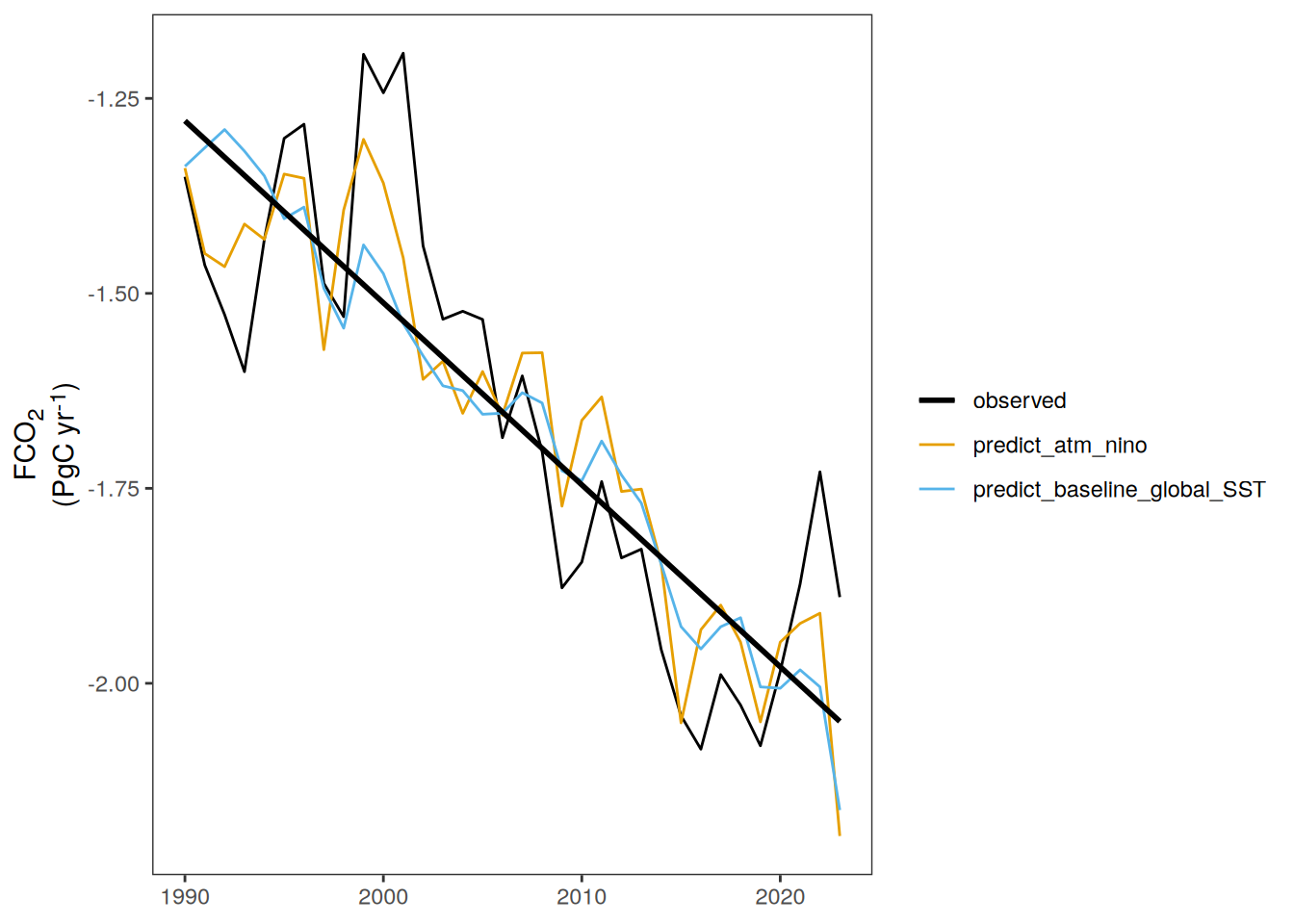
ggsave(width = 8,
height = 4,
dpi = 600,
filename = "../output/timeseries_pco2_products_predictions.jpg")
mlr %>%
mutate(
unexpected_baseline_global_SST =
fgco2_int_observed - fgco2_int_predict_baseline_global_SST,
unexpected_atm_nino =
fgco2_int_observed - fgco2_int_predict_atm_nino,
) %>%
select(product, year, starts_with("unex")) %>%
pivot_longer(starts_with("unex"),
names_prefix = "unexpected_") %>%
ggplot(aes(year, value, col = name)) +
geom_hline(yintercept = 0) +
geom_path() +
scale_color_okabeito() +
facet_wrap(~ product) +
labs(y = labels_breaks("fgco2_int")$i_legend_title) +
theme(axis.title.x = element_blank(),
axis.title.y = element_markdown(),
legend.title = element_blank())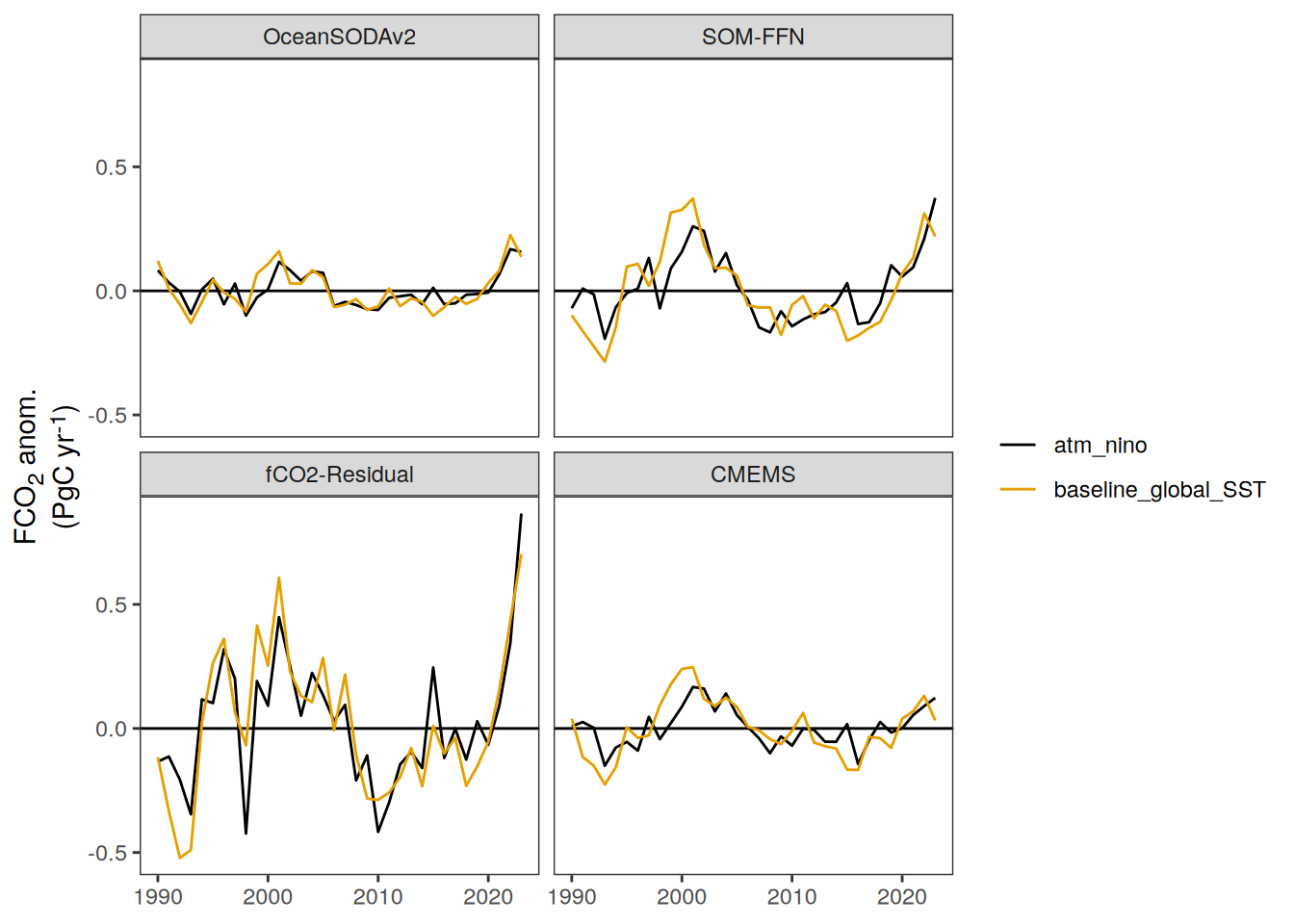
| Version | Author | Date |
|---|---|---|
| b031240 | jens-daniel-mueller | 2025-03-11 |
mlr_ensemble <- mlr %>%
mutate(
unexpected_baseline_global_SST =
fgco2_int_observed - fgco2_int_predict_baseline_global_SST,
unexpected_atm_nino =
fgco2_int_observed - fgco2_int_predict_atm_nino,
) %>%
select(product, year, starts_with("unex")) %>%
pivot_longer(starts_with("unex"), names_prefix = "unexpected_") %>%
group_by(year, name) %>%
summarise(sd = sd(value),
value = mean(value)) %>%
ungroup()
mlr_ensemble %>%
ggplot(aes(year, value,
ymin = value - sd,
ymax = value + sd)) +
geom_hline(yintercept = 0) +
geom_ribbon(alpha = 0.2) +
geom_path() +
scale_color_okabeito() +
facet_wrap(~ name) +
labs(y = labels_breaks("fgco2_int")$i_legend_title) +
theme(axis.title.x = element_blank(),
axis.title.y = element_markdown(),
legend.title = element_blank())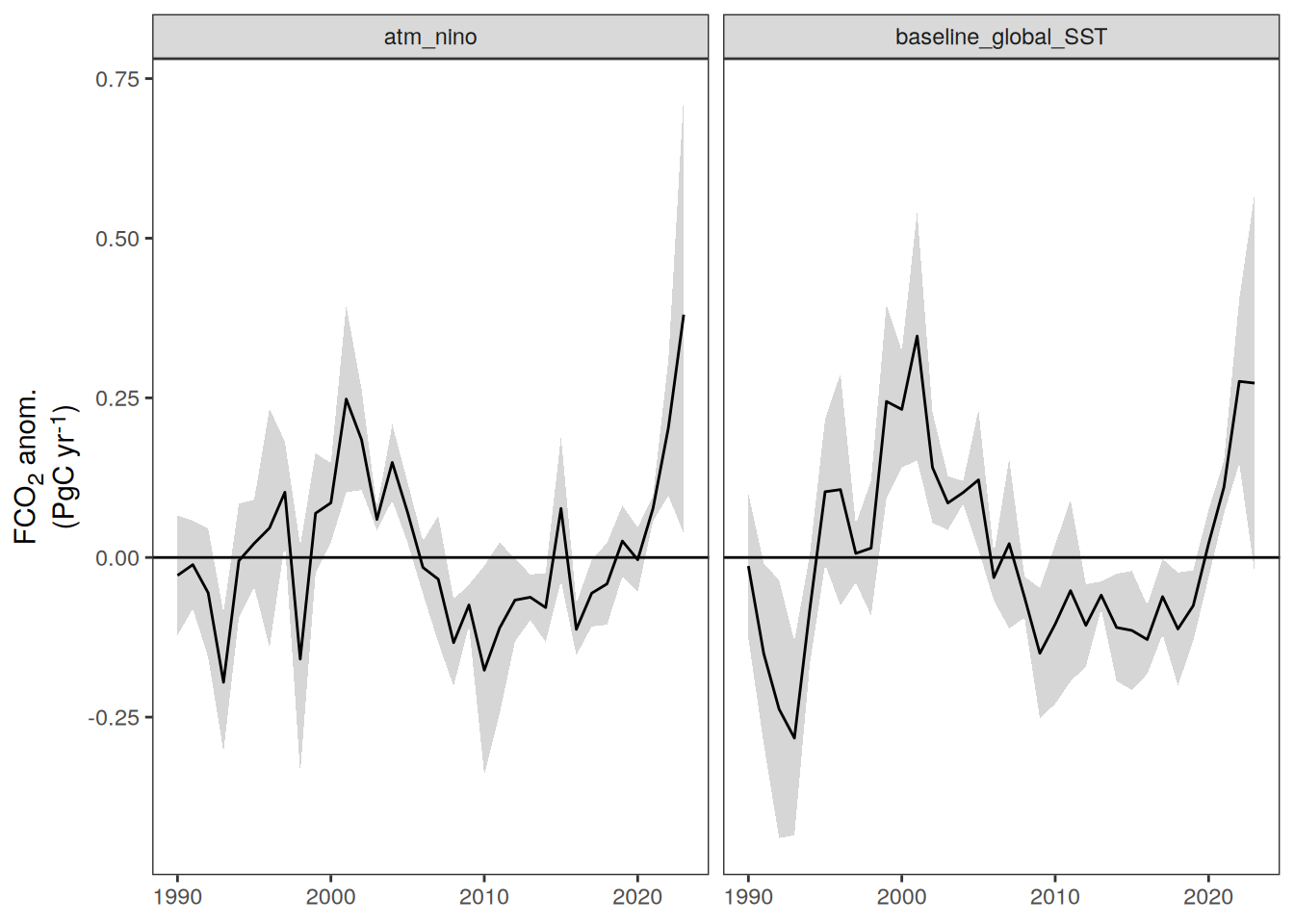
| Version | Author | Date |
|---|---|---|
| b031240 | jens-daniel-mueller | 2025-03-11 |
ggsave(width = 8,
height = 4,
dpi = 600,
filename = "../output/timeseries_pco2_products_predictions_ensemble_mean.jpg")
mlr_ensemble %>%
filter(year == 2023)# A tibble: 2 × 4
year name sd value
<dbl> <chr> <dbl> <dbl>
1 2023 atm_nino 0.343 0.380
2 2023 baseline_global_SST 0.297 0.273pco2_product_biome_annual_anomaly %>%
filter(year == 2023,
name %in% c("fgco2", "fgco2_int", "dfco2",
"kw_sol", "temperature",
"no3", "mld", "intpp", "chl")) %>%
mutate(region = case_when(biome == "Global non-polar" ~ "Global non-polar",
# biome %in% super_biomes ~ "Super biomes",
TRUE ~ "Biomes"),
region = factor(region, levels = c("Global non-polar", "Biomes"))) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_col(aes(biome, value, fill = product),
position = "dodge2") +
scale_fill_manual(values = color_products) +
geom_col(aes(biome, fit, group = product, col = paste0(2023,"\nlinear\nprediction")),
position = "dodge2",
fill = "transparent") +
labs(y = labels_breaks(unique(.x$name))$i_legend_title,
title = "Absolute") +
scale_color_grey() +
facet_grid(.~region, scales = "free_x", space = "free_x") +
theme(legend.title = element_blank(),
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1),
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
strip.background = element_blank(),
legend.position = "top")
)[[1]]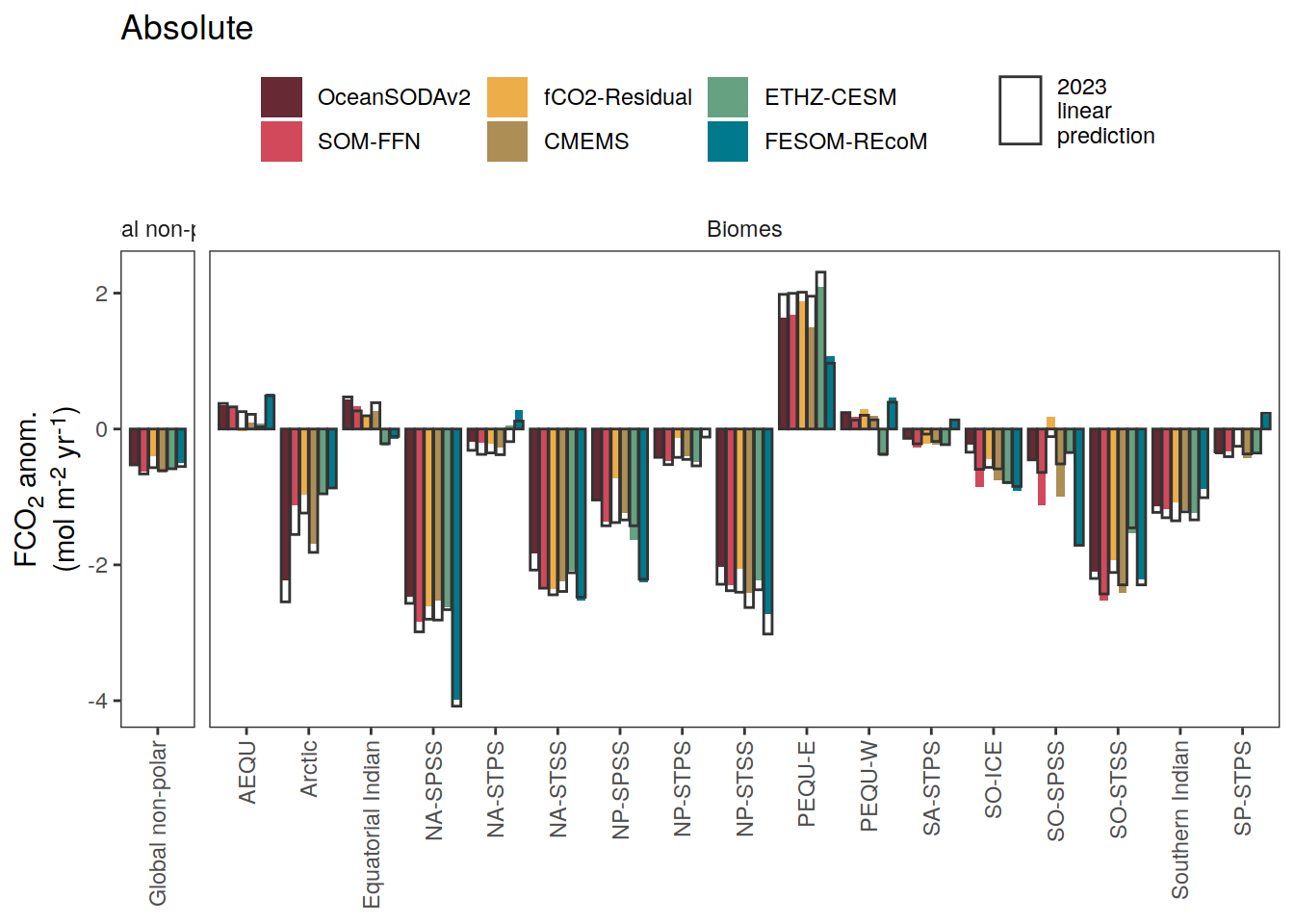
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[2]]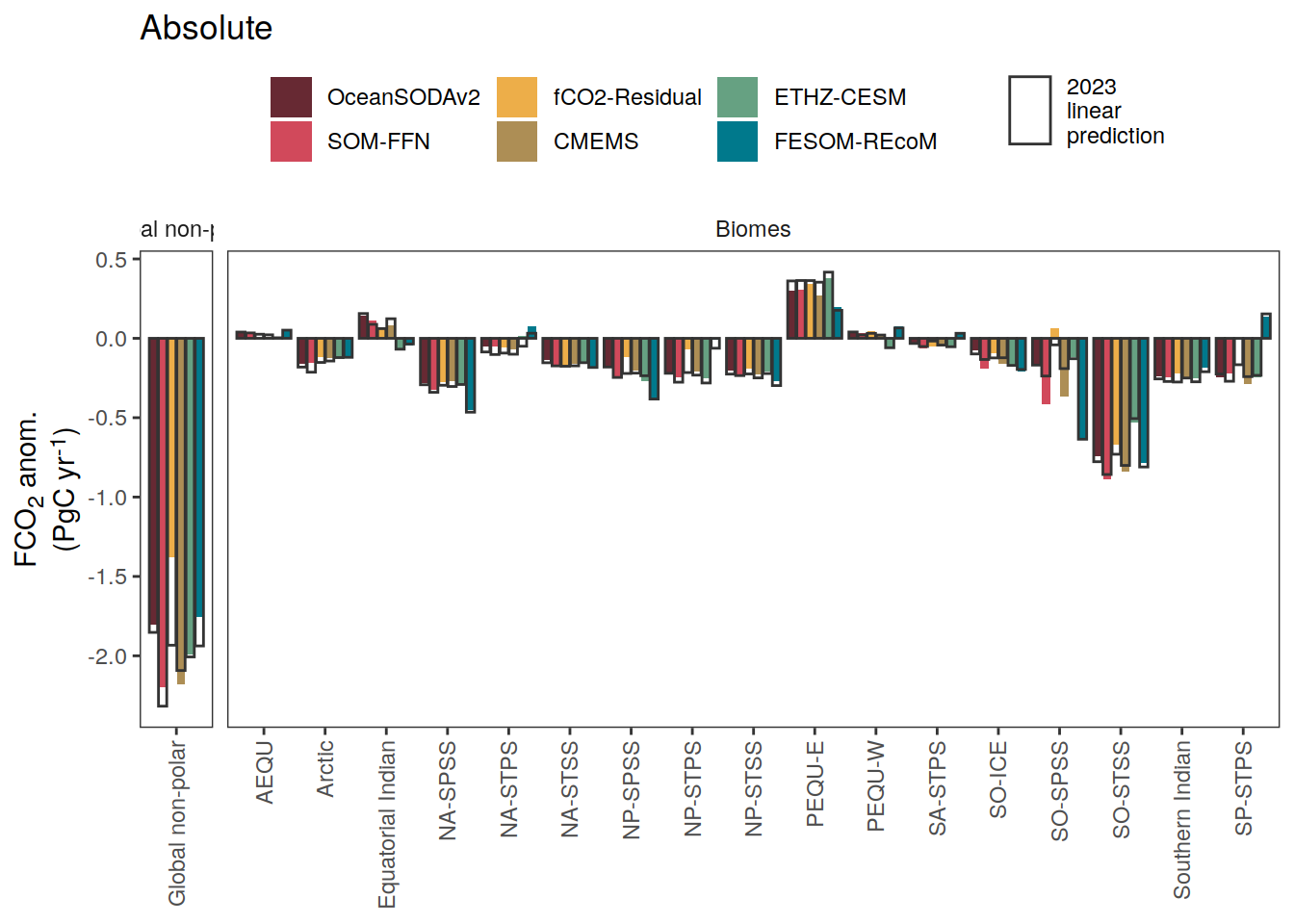
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[3]]
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[4]]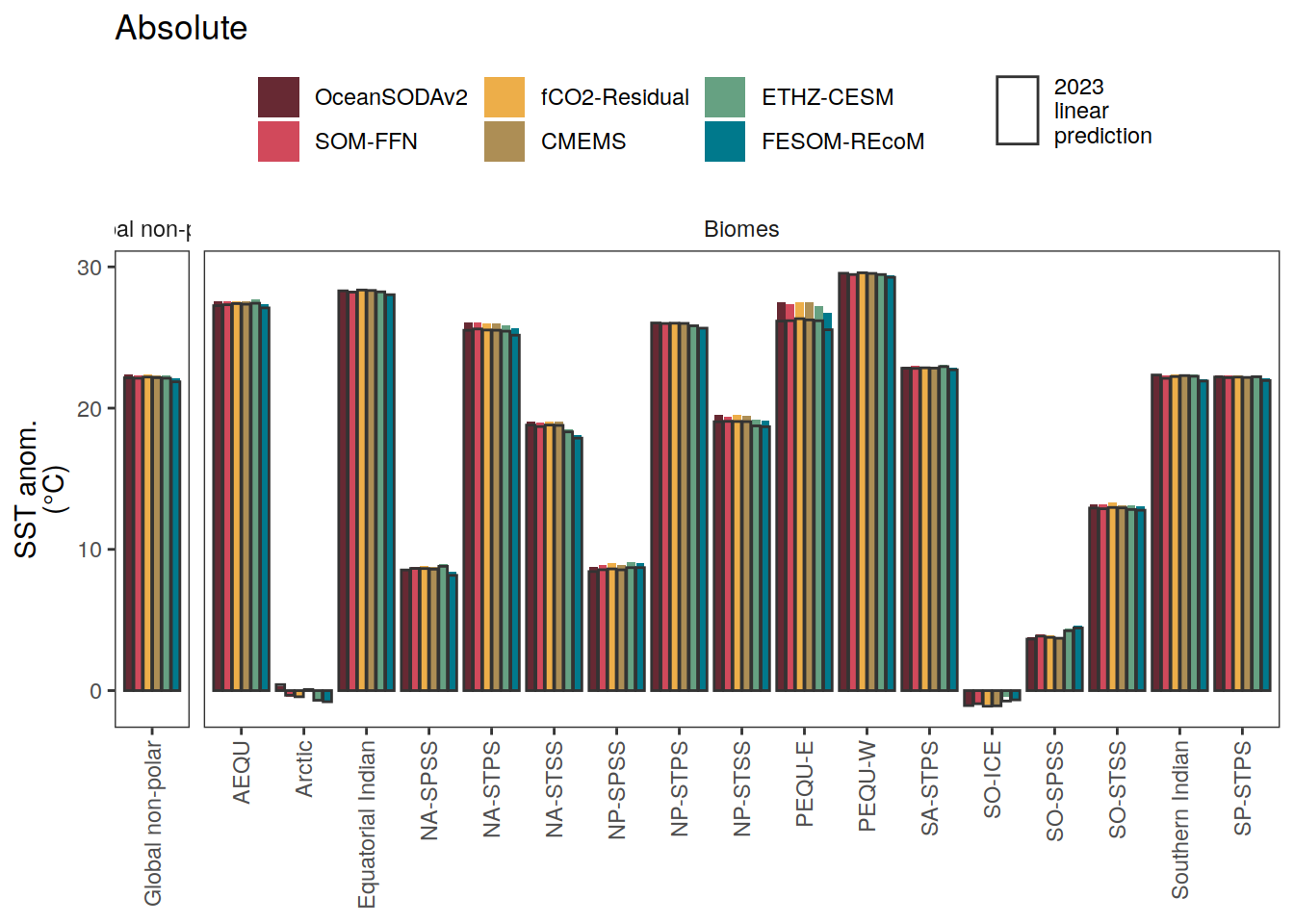
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[5]]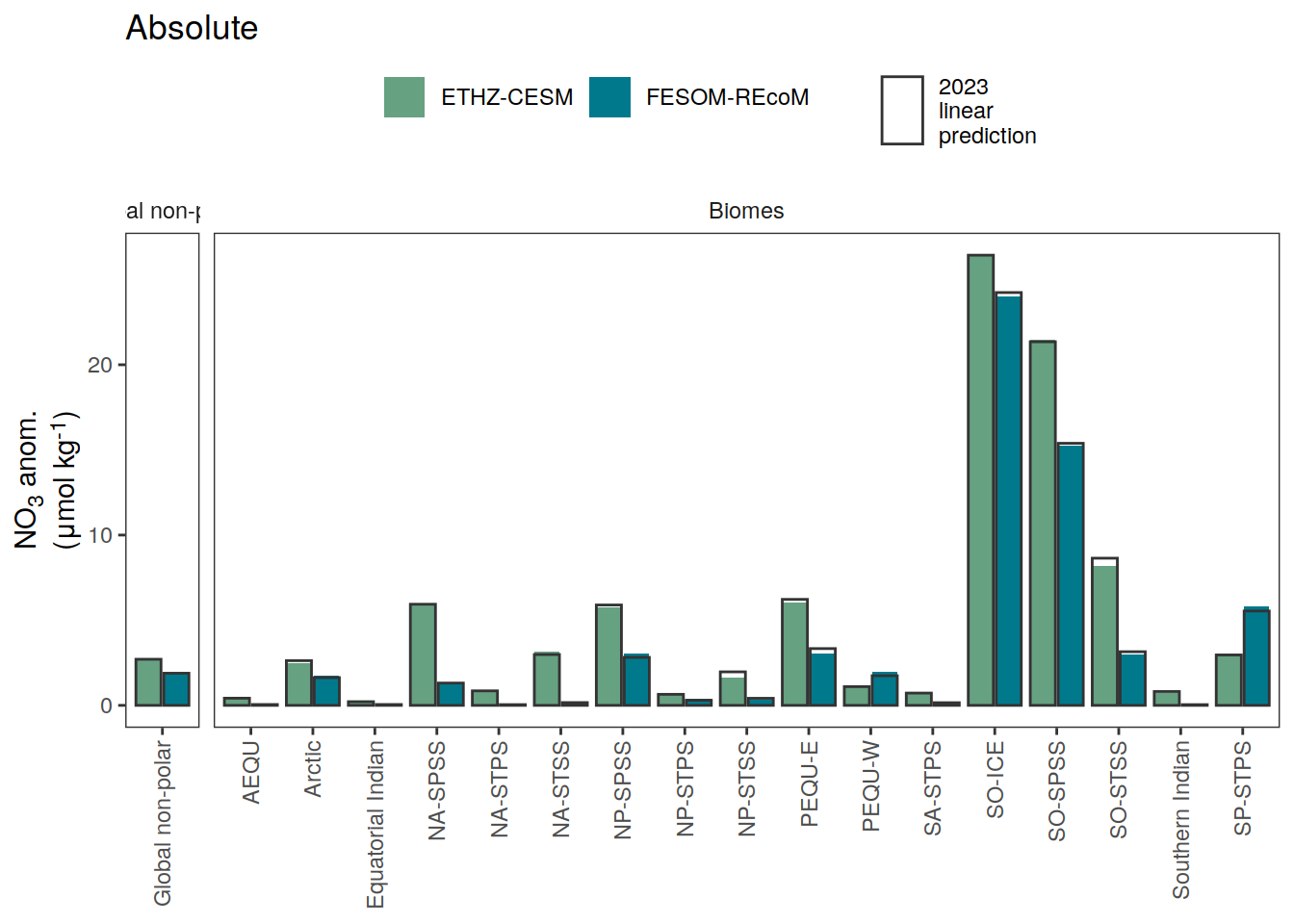
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[6]]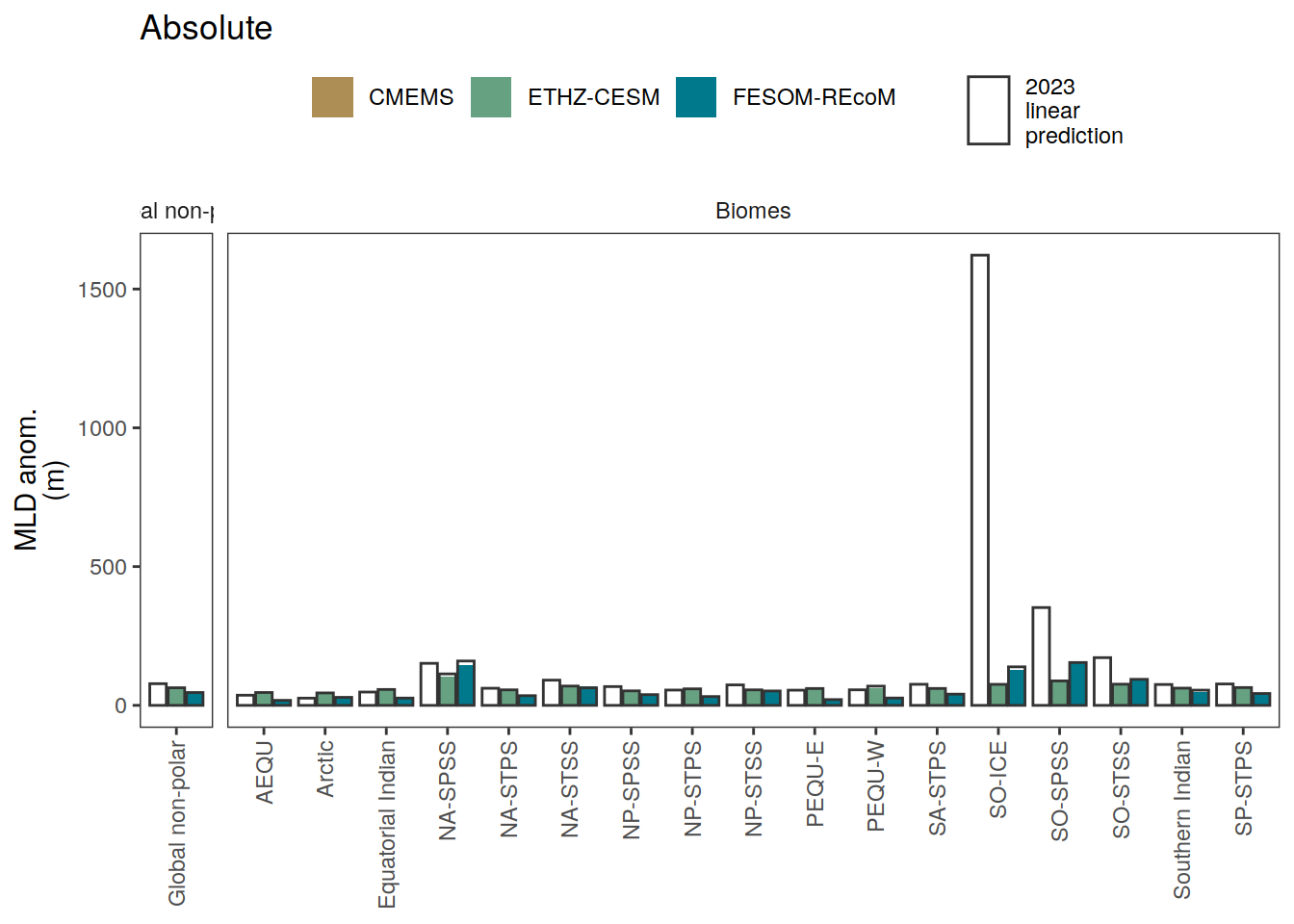
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[7]]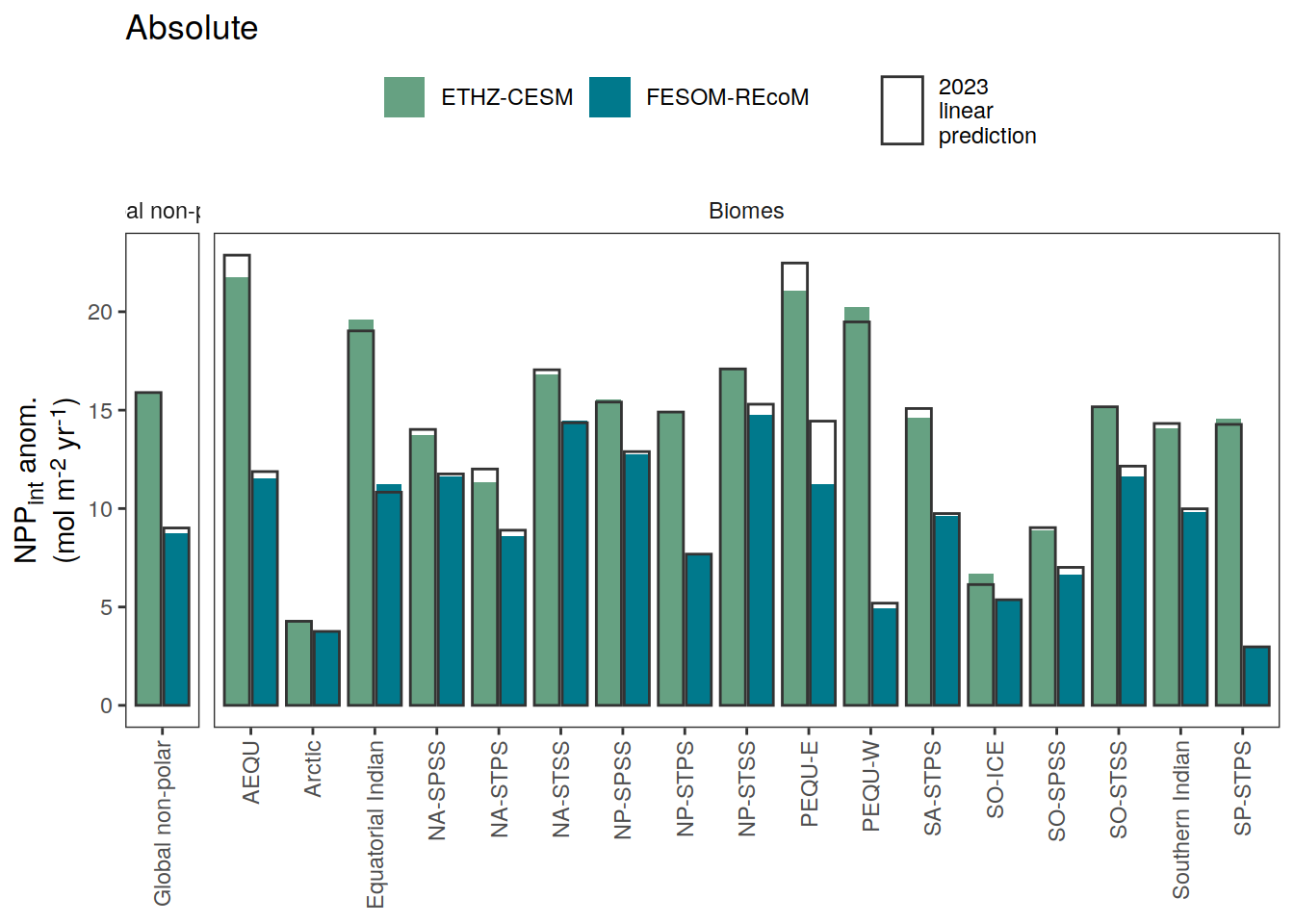
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[8]]
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2a28b07 | jens-daniel-mueller | 2024-07-22 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 7f9c687 | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
full_join(
pco2_product_biome_annual_anomaly %>%
filter(year != 2023,
name %in% name_core) %>%
group_by(product, name, biome) %>%
summarise(resid_sd = sd(resid)) %>%
ungroup(),
pco2_product_biome_annual_anomaly %>%
filter(year == 2023,
name %in% name_core)) %>%
mutate(
region = case_when(
biome == "Global non-polar" ~ "Global non-polar",
TRUE ~ "Biomes"
),
region = factor(region, levels = c("Global non-polar", "Biomes"))
) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_col(aes(biome, value - fit, fill = product),
position = "dodge2") +
scale_fill_manual(values = color_products) +
geom_col(aes(biome, resid_sd * sign(value - fit),
group = product, col = paste0("Anomaly SD\nexcl.",2023)),
position = "dodge2",
fill = "transparent") +
labs(y = labels_breaks(unique(.x$name))$i_legend_title,
title = "Anomalies") +
scale_color_grey() +
facet_grid(.~region, scales = "free_x", space = "free_x") +
theme(legend.title = element_blank(),
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1),
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
strip.background = element_blank(),
legend.position = "top")
)[[1]]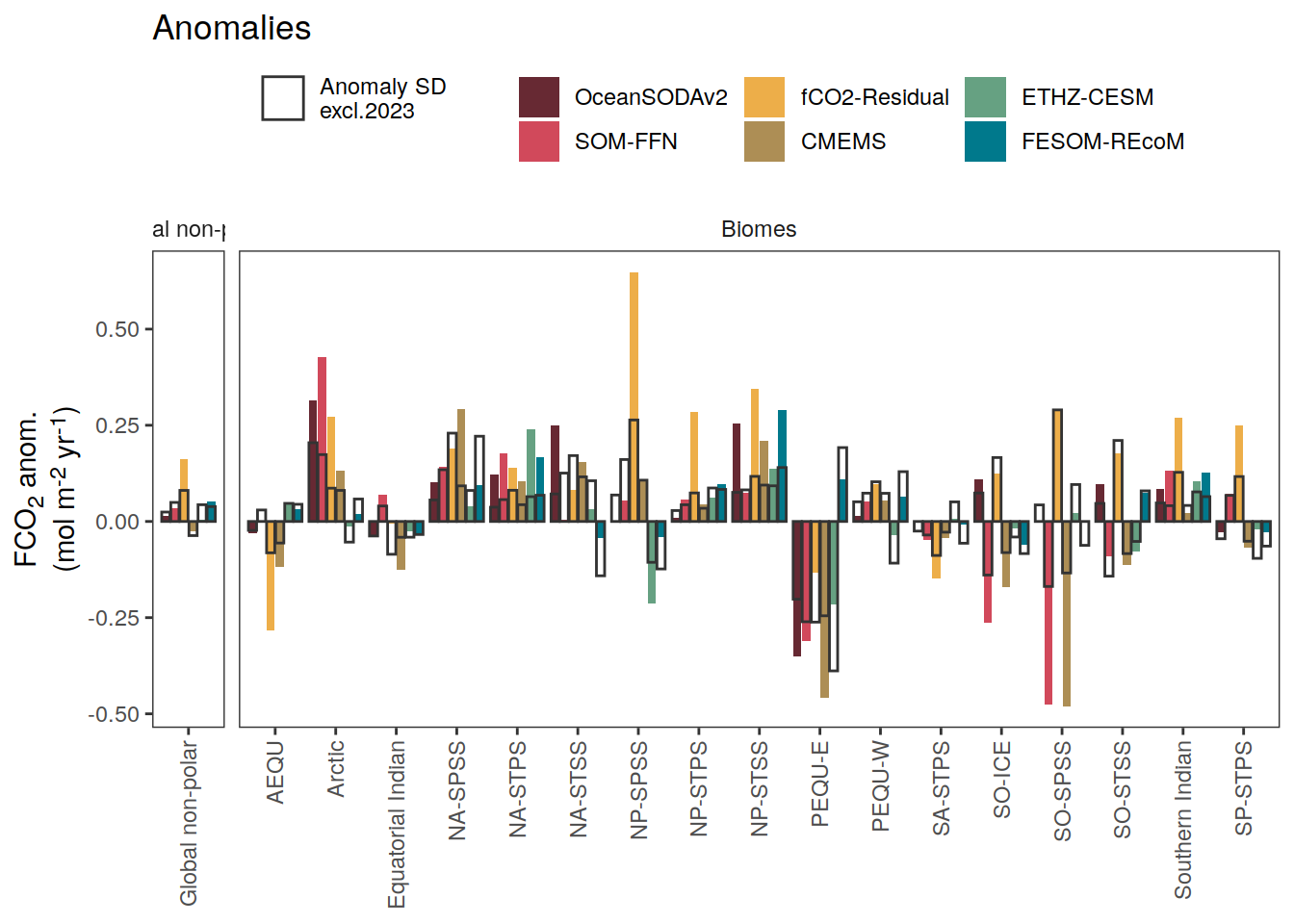
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[2]]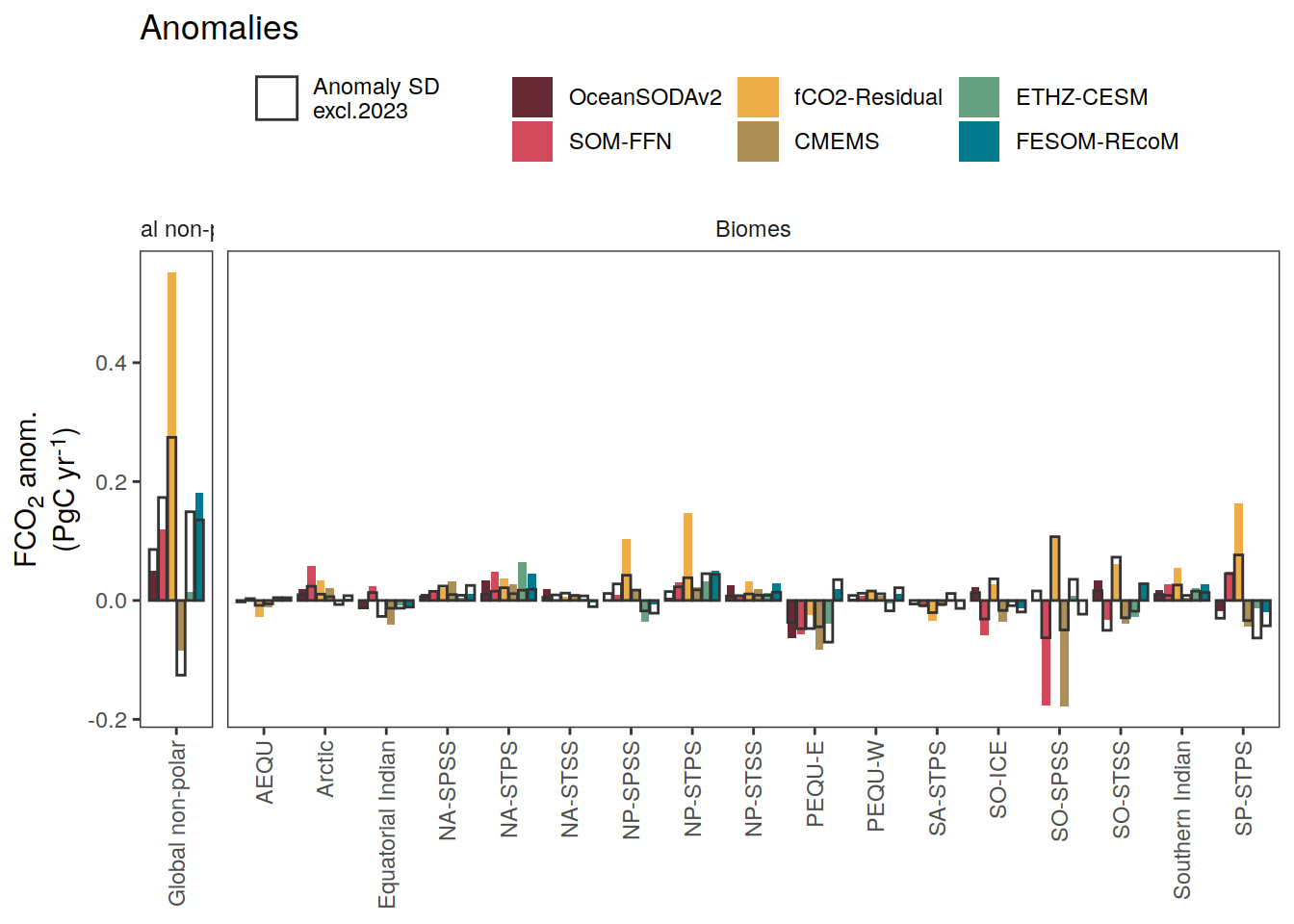
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[3]]
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[4]]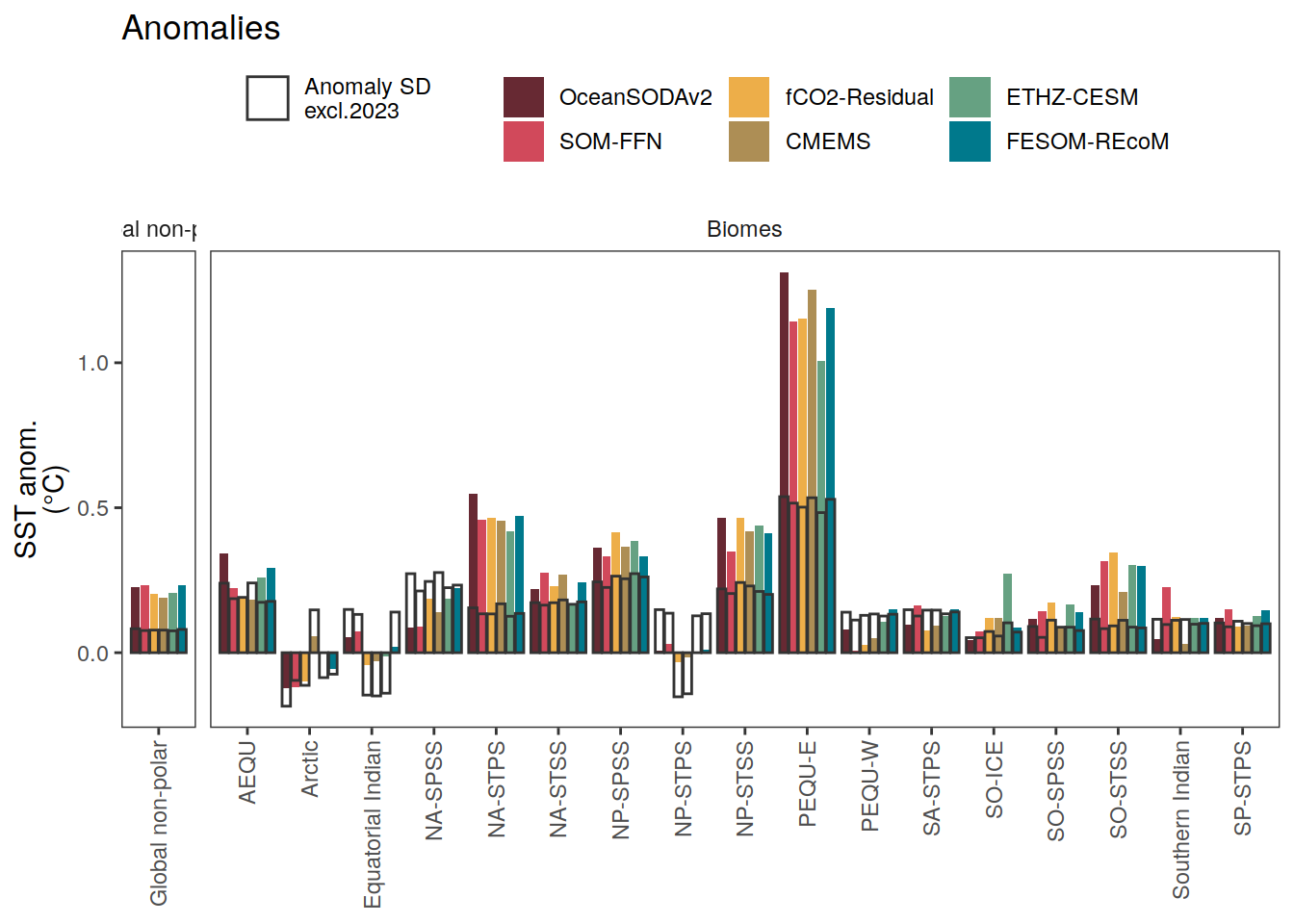
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[5]]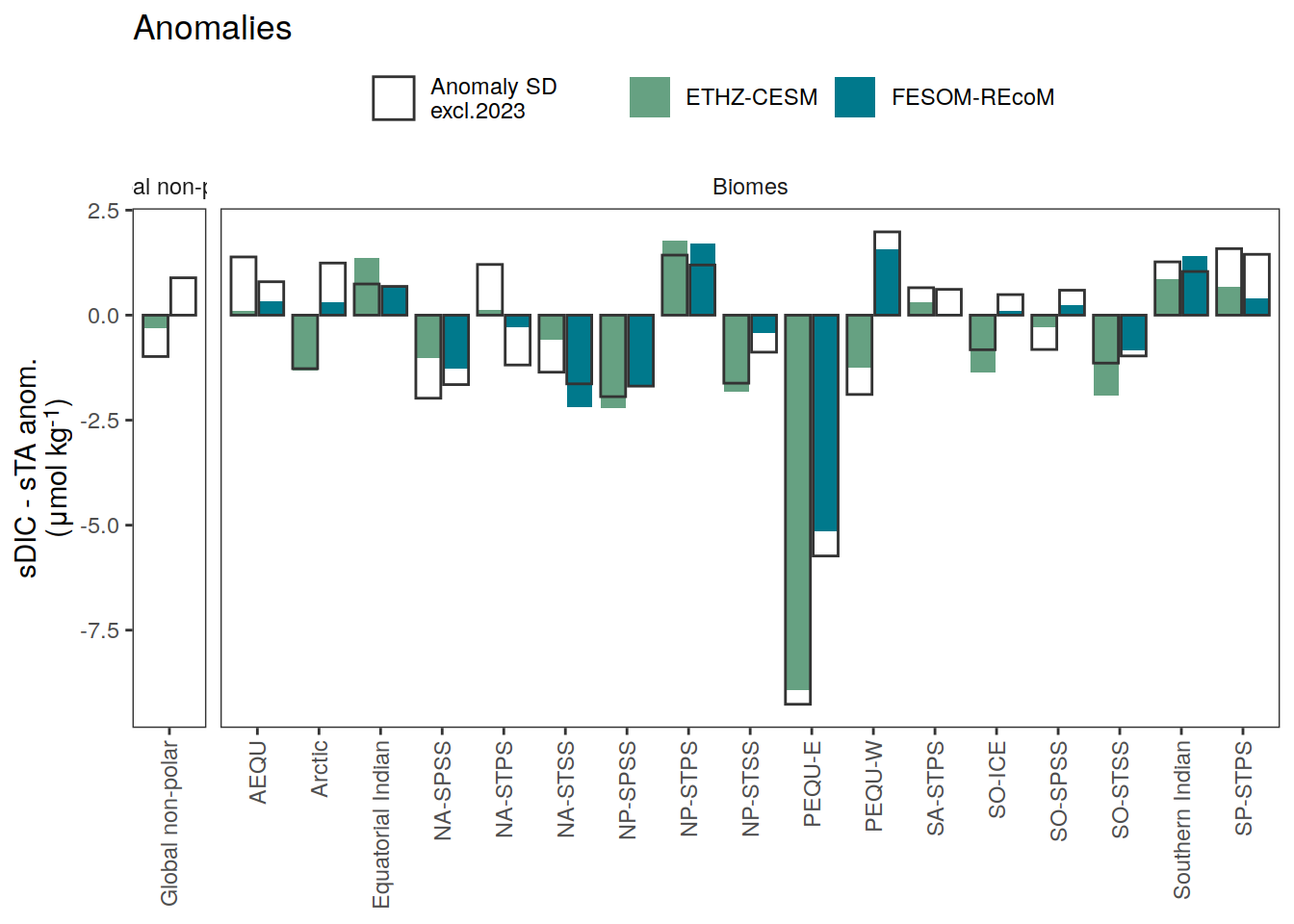
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[6]]
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[7]]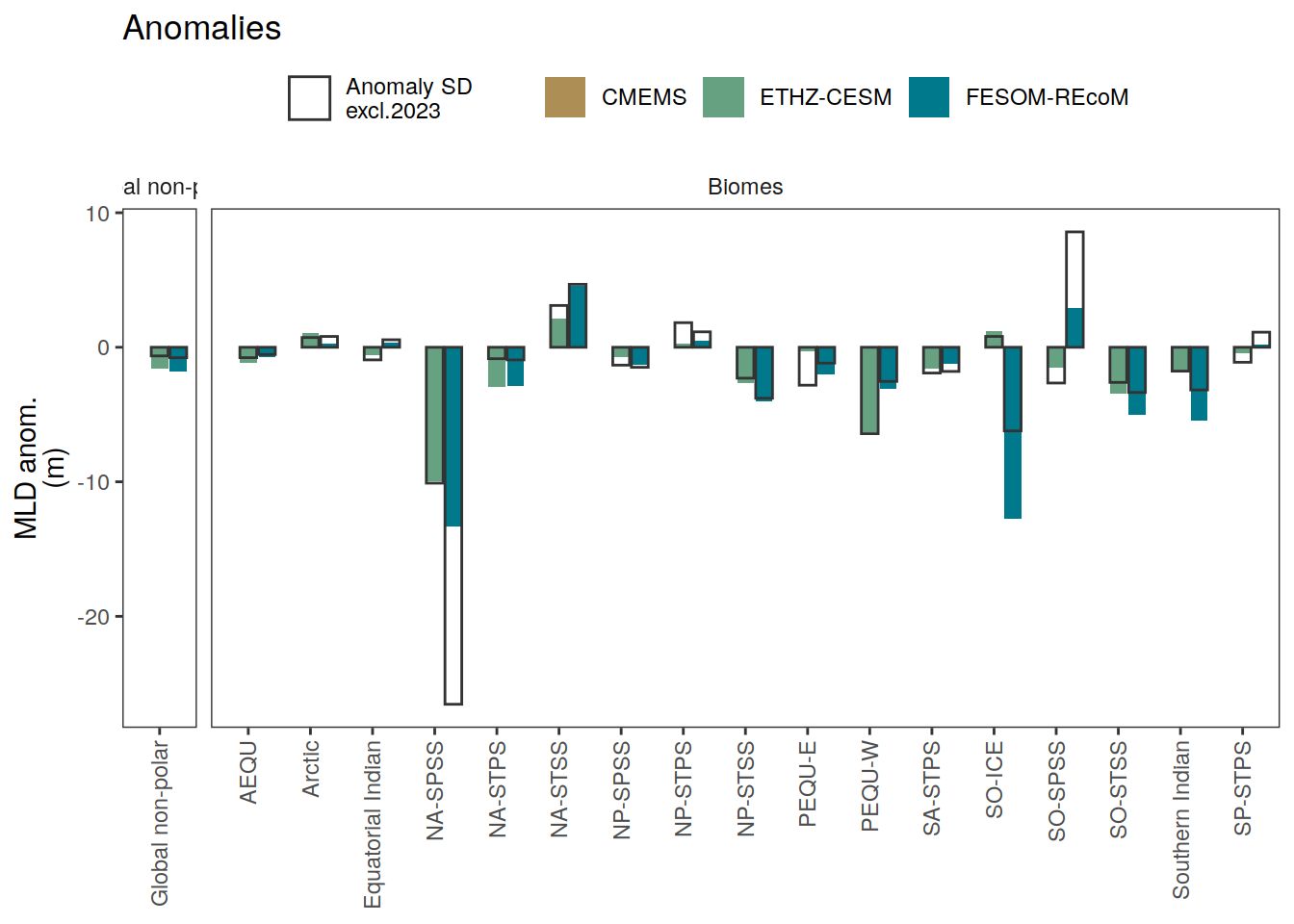
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[8]]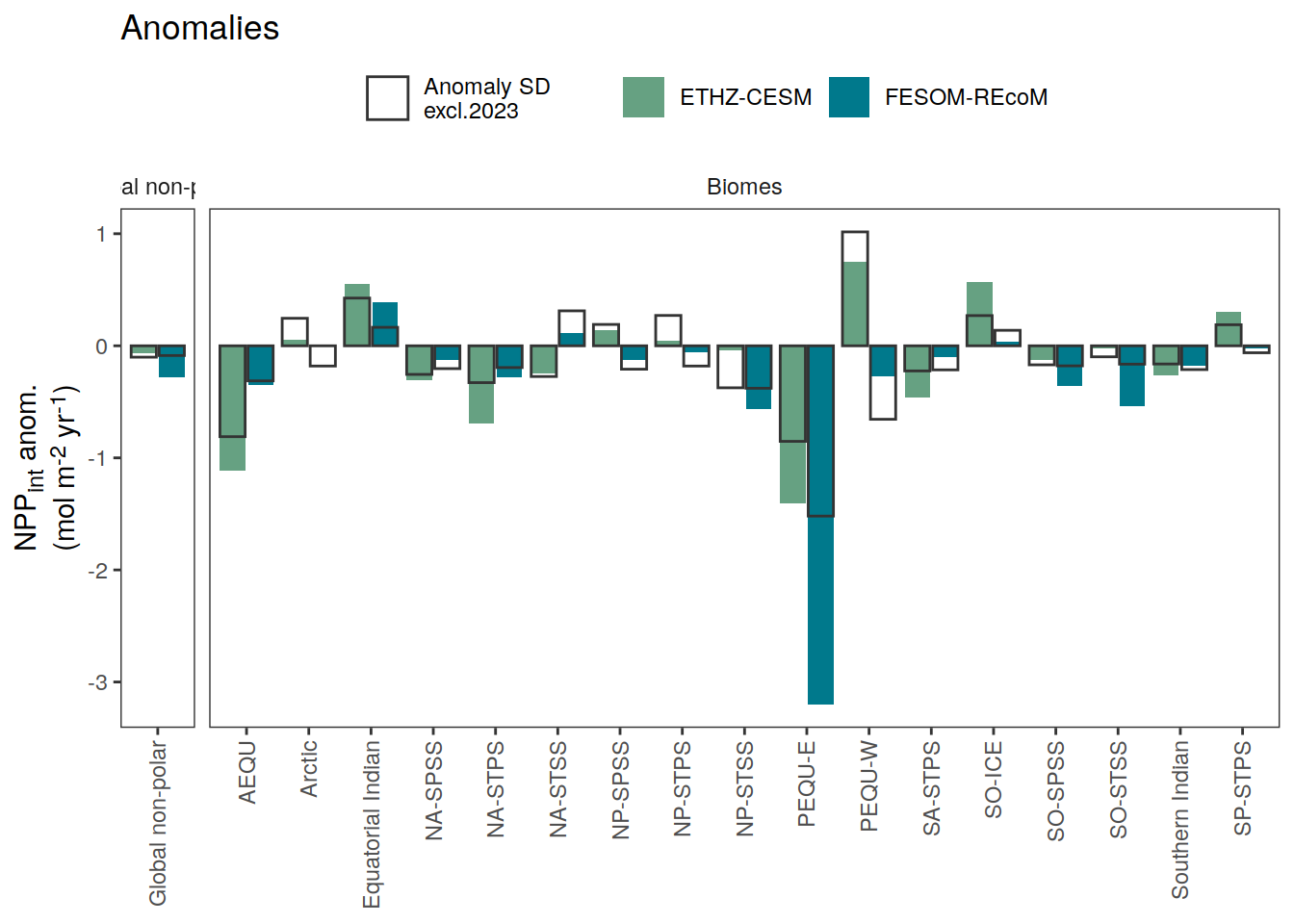
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 7f9c687 | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
[[9]]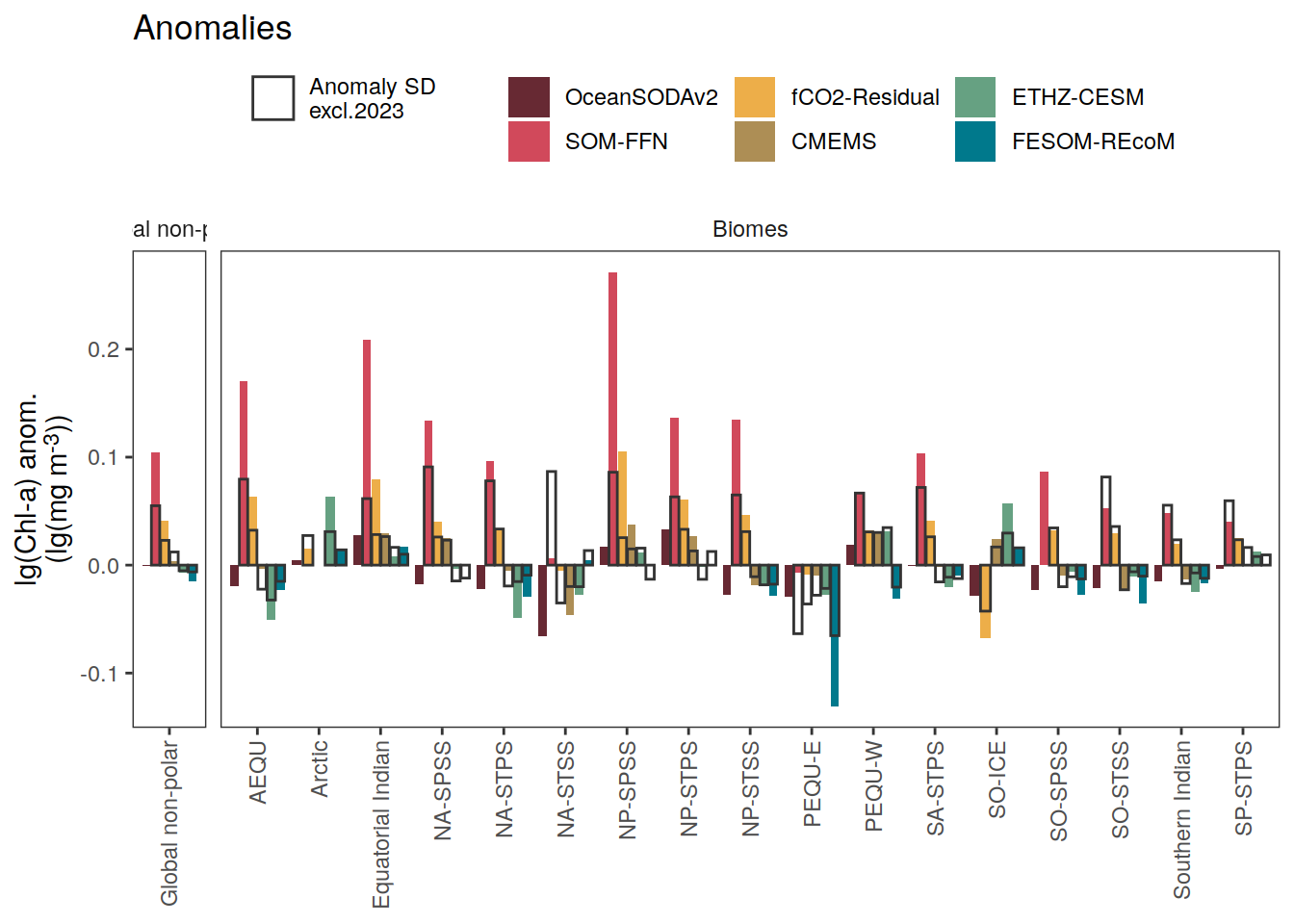
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
Super regions
pco2_product_biome_annual_anomaly_super_regions <-
full_join(
pco2_product_biome_annual_anomaly %>%
filter(biome != "Global non-polar"),
biome_mask %>%
mutate(area = earth_surf(lat, lon)) %>%
group_by(biome) %>%
summarise(area = sum(area)) %>%
ungroup()
) %>%
pivot_longer(c(value,resid,fit),
names_to = "estimate") %>%
pivot_wider()
pco2_product_biome_annual_anomaly_super_regions <-
bind_rows(
pco2_product_biome_annual_anomaly_super_regions %>%
select(-biome) %>%
group_by(product, estimate, year) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(., na.rm = TRUE))) %>%
ungroup() %>%
mutate(region = "Global"),
pco2_product_biome_annual_anomaly_super_regions %>%
filter(!str_detect(biome, "SO-ICE|SO-SPSS|Arctic")) %>%
select(-biome) %>%
group_by(product, estimate, year) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(., na.rm = TRUE))) %>%
ungroup() %>%
mutate(region = "Global non-polar"),
pco2_product_biome_annual_anomaly_super_regions %>%
filter(!str_detect(biome, "SO-ICE|Arctic")) %>%
select(-biome) %>%
group_by(product, estimate, year) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(., na.rm = TRUE))) %>%
ungroup() %>%
mutate(region = "Global ice-free"),
pco2_product_biome_annual_anomaly_super_regions %>%
filter(str_detect(biome, "NA-|NP-")) %>%
select(-biome) %>%
group_by(product, estimate, year) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(., na.rm = TRUE))) %>%
ungroup() %>%
mutate(region = "NH extratropics"),
# pco2_product_biome_annual_anomaly_super_regions %>%
# filter(str_detect(biome, "NA-")) %>%
# select(-biome) %>%
# group_by(product, estimate, year) %>%
# summarise(across(-c(fgco2_int, area),
# ~ weighted.mean(., area, na.rm = TRUE)),
# across(fgco2_int,
# ~ sum(., na.rm = TRUE))) %>%
# ungroup() %>%
# mutate(region = "North Atlantic"),
# pco2_product_biome_annual_anomaly_super_regions %>%
# filter(str_detect(biome, "NP-")) %>%
# select(-biome) %>%
# group_by(product, estimate, year) %>%
# summarise(across(-c(fgco2_int, area),
# ~ weighted.mean(., area, na.rm = TRUE)),
# across(fgco2_int,
# ~ sum(., na.rm = TRUE))) %>%
# ungroup() %>%
# mutate(region = "North Pacific"),
pco2_product_biome_annual_anomaly_super_regions %>%
filter(str_detect(biome, "PEQU|AEQU|Equ")) %>%
select(-biome) %>%
group_by(product, estimate, year) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(., na.rm = TRUE))) %>%
ungroup() %>%
mutate(region = "Tropics"),
pco2_product_biome_annual_anomaly_super_regions %>%
filter(str_detect(biome, "SA-|SP-|Southern|SO-STSS")) %>%
select(-biome) %>%
group_by(product, estimate, year) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(., na.rm = TRUE))) %>%
ungroup() %>%
mutate(region = "SH extratropics")) %>%
mutate(region = fct_inorder(region)) %>%
pivot_longer(-c(product, year, region, estimate)) %>%
pivot_wider(names_from = estimate)
pco2_product_biome_annual_anomaly_super_regions %>%
filter(year == 2023,
name %in% c("fgco2", "fgco2_int", "dfco2", "temperature")) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_hline(yintercept = 0) +
geom_col(aes(region, value, fill = product),
position = "dodge2") +
scale_fill_manual(values = color_products) +
geom_col(aes(region, fit, group = product, col = paste0(2023,"\nlinear\nprediction")),
position = "dodge2",
fill = "transparent") +
labs(y = str_remove(labels_breaks(unique(.x$name))$i_legend_title, " anom.")) +
scale_color_grey() +
facet_grid(.~region, scales = "free_x", space = "free_x") +
theme(legend.title = element_blank(),
axis.text.x = element_blank(),
axis.title.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.y = element_markdown(),
strip.background = element_blank(),
legend.position = "top")
)[[1]]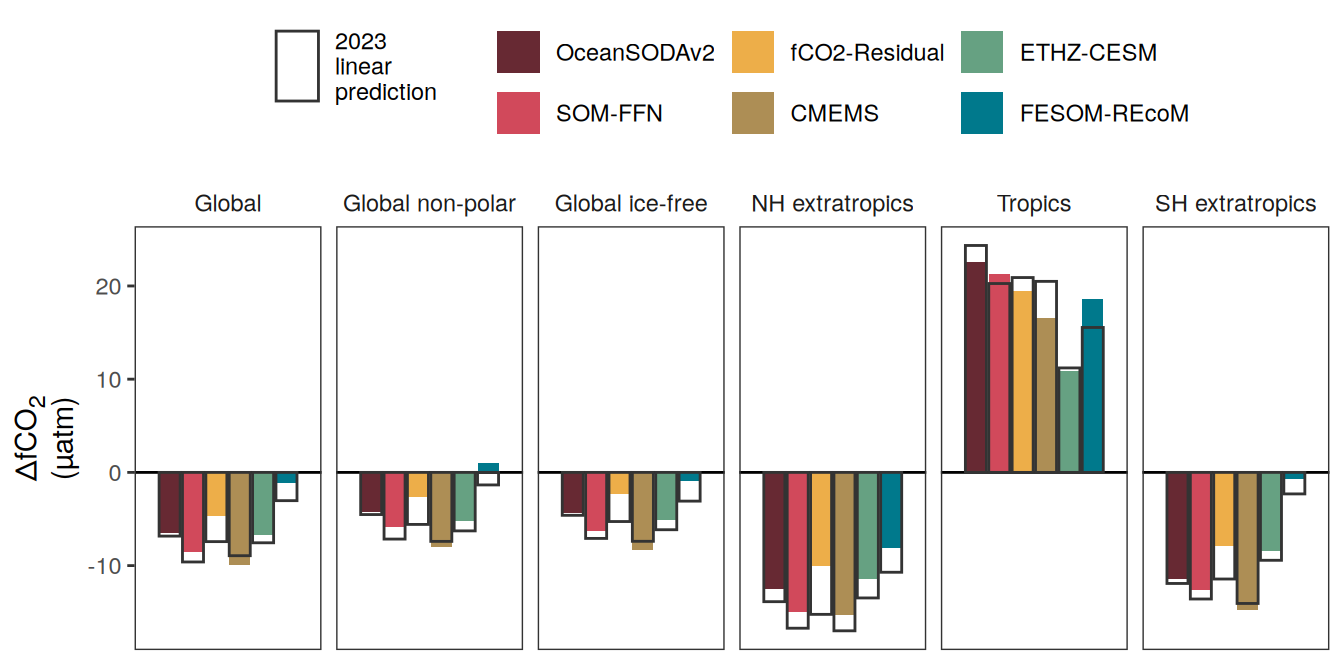
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| 6a96e1f | jens-daniel-mueller | 2024-08-26 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4897f6e | jens-daniel-mueller | 2024-07-08 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
[[2]]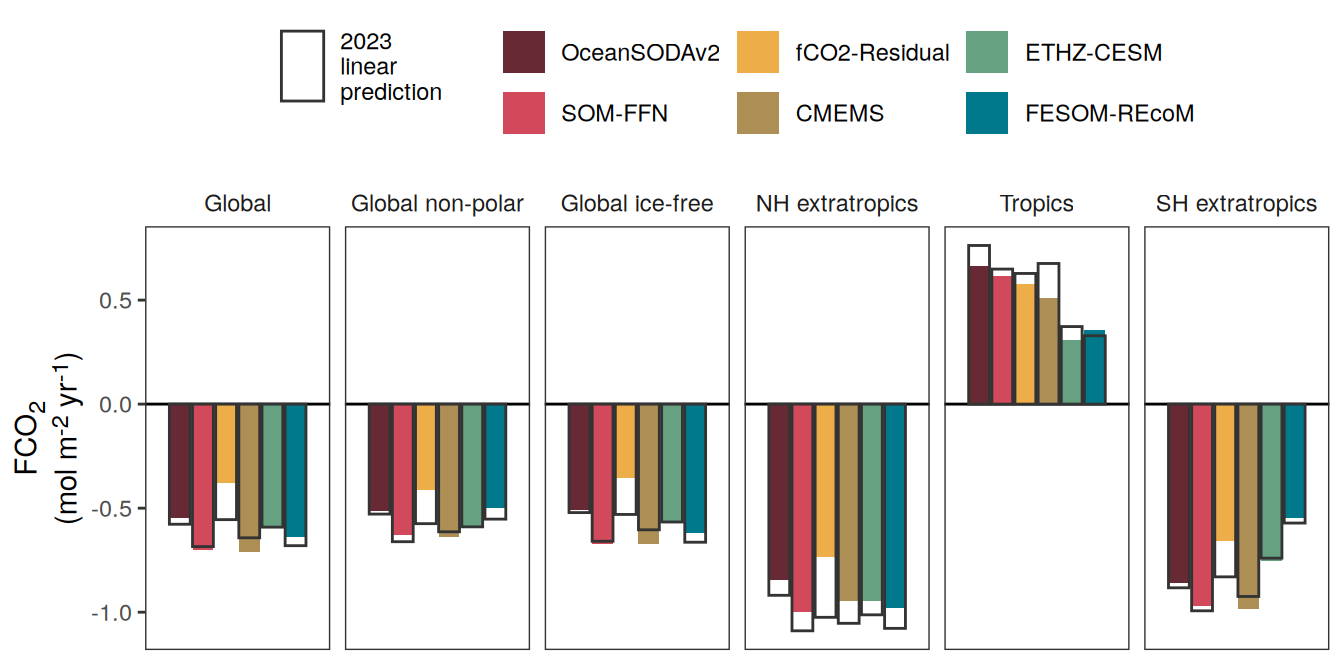
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
[[3]]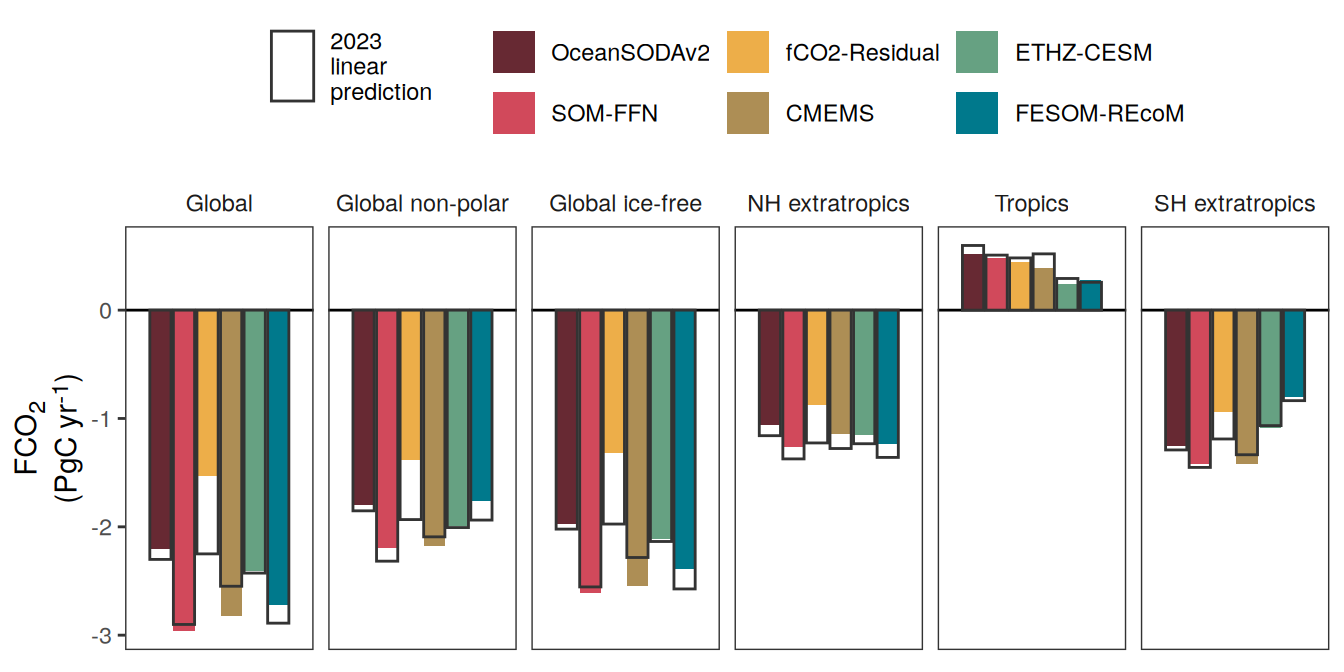
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
[[4]]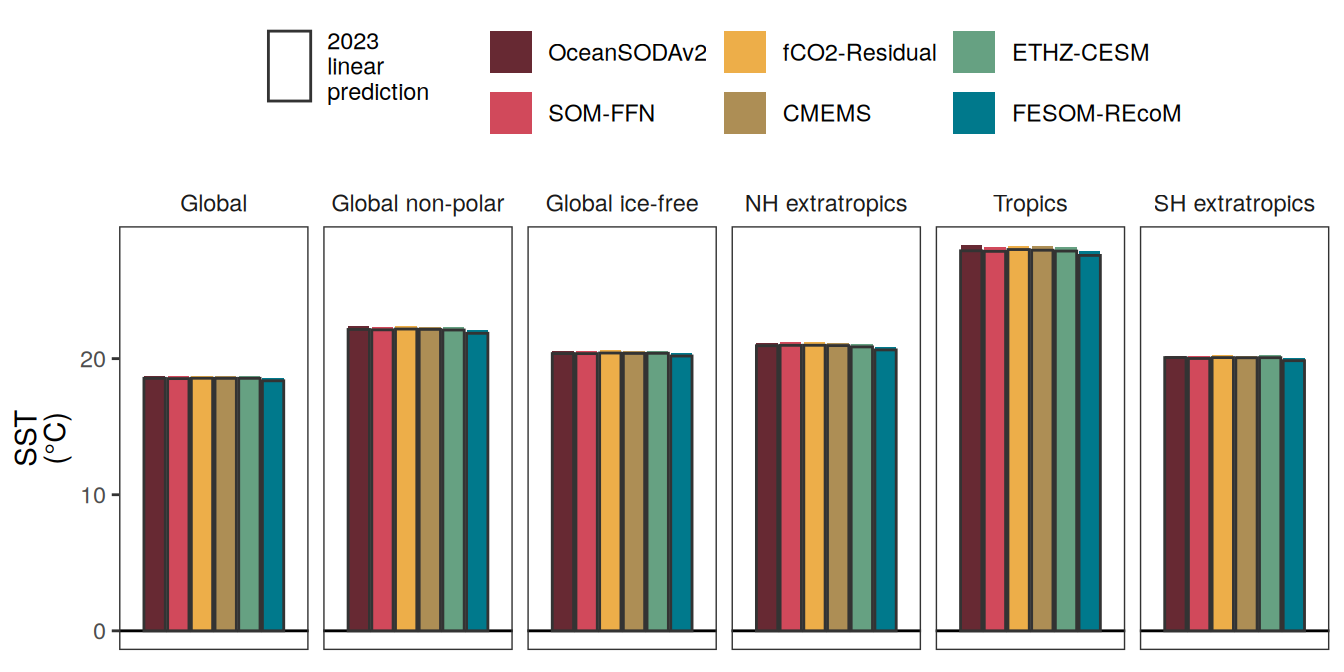
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
full_join(pco2_product_biome_annual_anomaly_super_regions %>%
group_by(product, name, region) %>%
summarise(resid_sd = sd(resid, na.rm = TRUE)) %>%
ungroup(),
pco2_product_biome_annual_anomaly_super_regions %>%
filter(year == 2023)) %>%
filter(name %in% c("fgco2", "fgco2_int", "dfco2", "temperature")) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_hline(yintercept = 0) +
geom_col(aes(region, resid, fill = product),
position = "dodge2") +
scale_fill_manual(values = color_products) +
geom_col(aes(region, resid_sd * sign(value - fit),
group = product, col = paste0("Anomaly SD\nexcl.",2023)),
position = "dodge2",
fill = "transparent") +
labs(y = labels_breaks(unique(.x$name))$i_legend_title) +
scale_color_grey() +
facet_grid(. ~ region, scales = "free_x", space = "free_x") +
theme(legend.title = element_blank(),
axis.text.x = element_blank(),
axis.title.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.y = element_markdown(),
strip.background = element_blank(),
legend.position = "top")
)[[1]]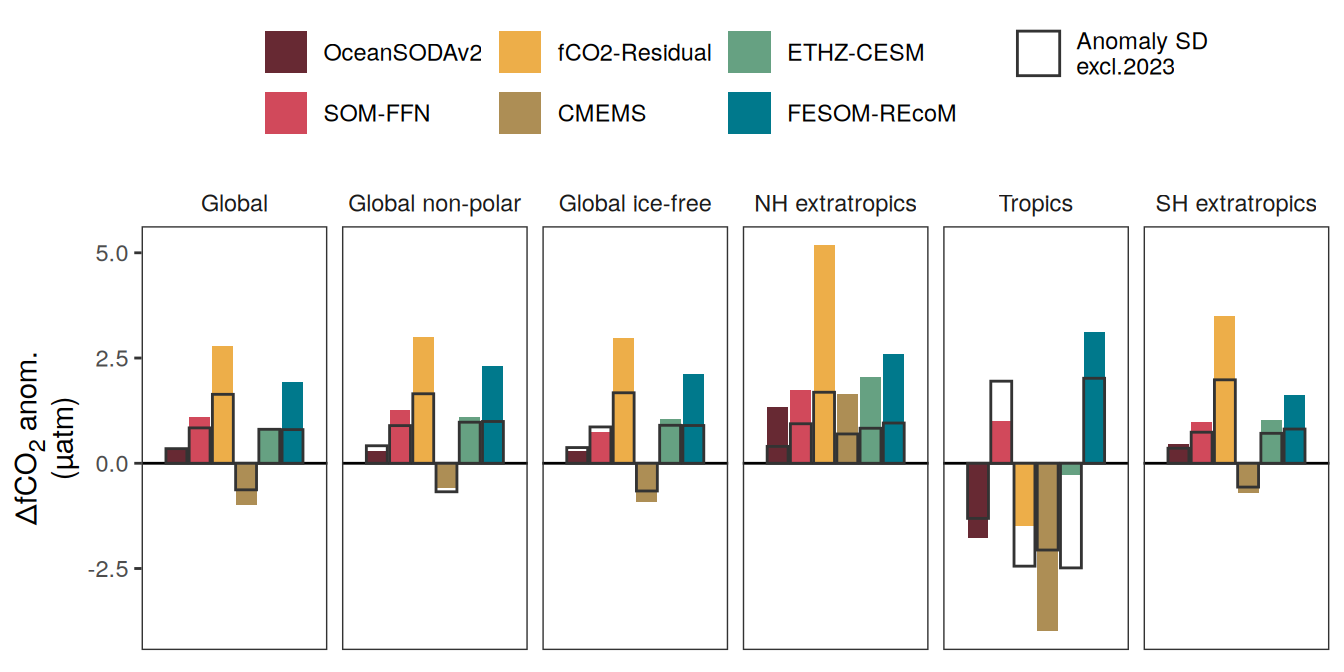
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
[[2]]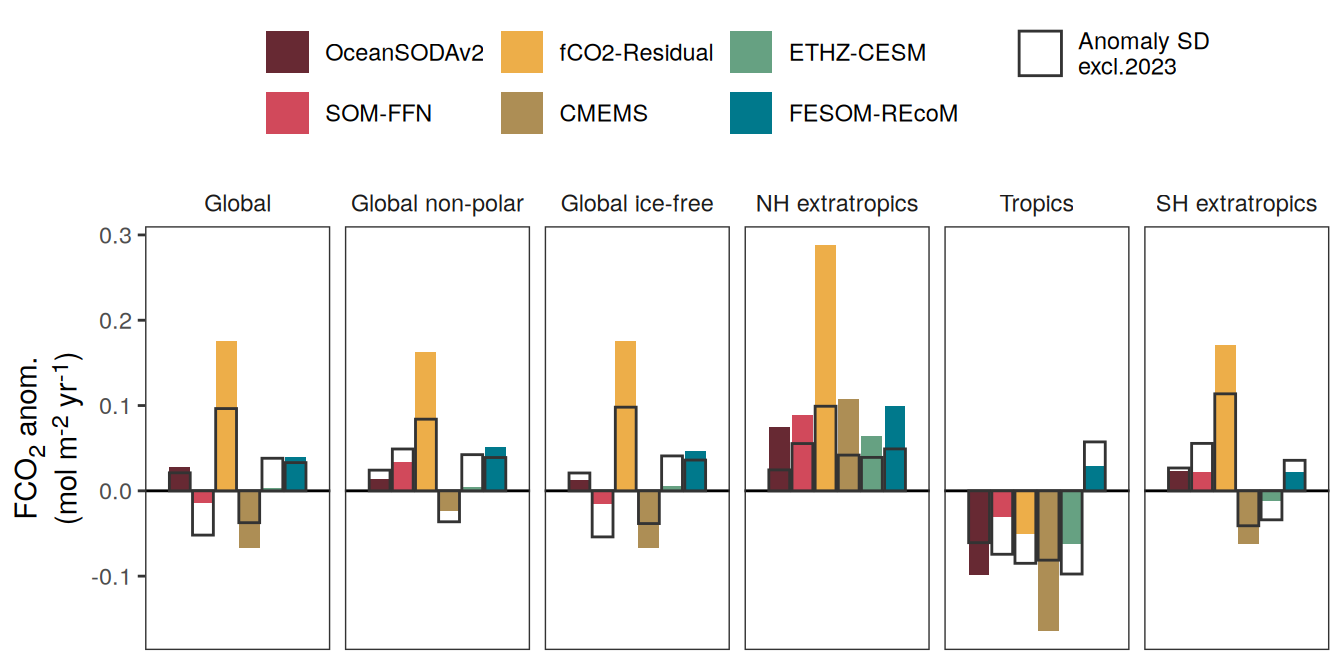
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
[[3]]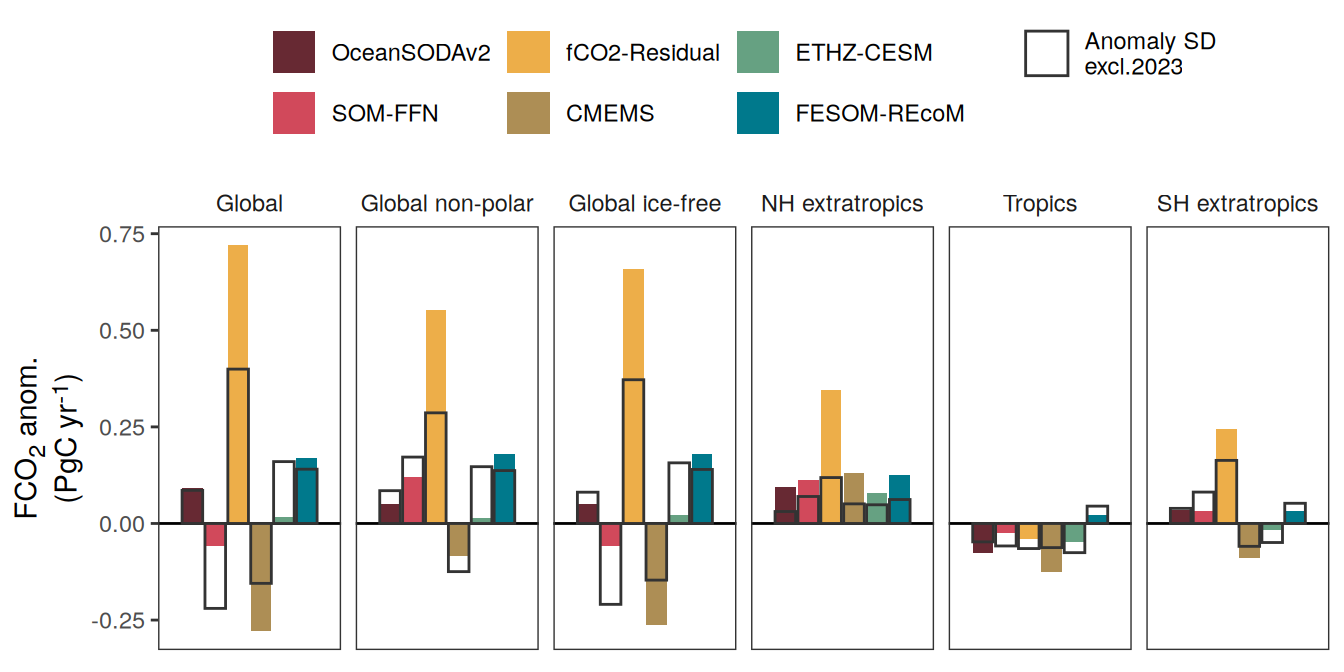
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
[[4]]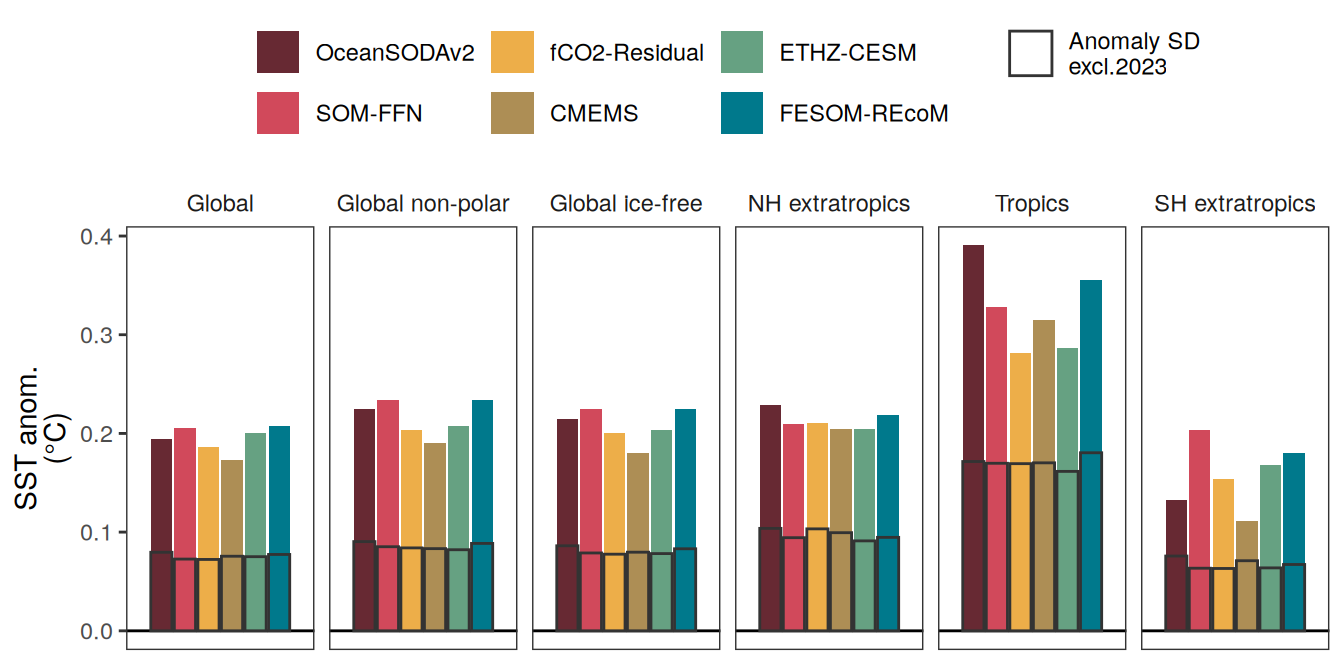
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 0f5b472 | jens-daniel-mueller | 2024-08-27 |
| 08ca8c7 | jens-daniel-mueller | 2024-08-27 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 7ad8576 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 4be90dd | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
pco2_product_biome_annual_anomaly_super_regions <-
bind_rows(
pco2_product_biome_annual_anomaly_super_regions %>%
rename(biome = region),
pco2_product_biome_annual_anomaly %>%
filter(biome != "Global non-polar")
) %>%
filter(product %in% pco2_product_list) %>%
group_by(year, name, biome) %>%
summarise(
resid_sd = sd(resid),
resid = mean(resid),
value_sd = sd(value),
value = mean(value)
) %>%
ungroup()
pco2_product_biome_annual_anomaly_super_regions <-
pco2_product_biome_annual_anomaly_super_regions %>%
filter(name %in% c("temperature", "fgco2", "fgco2_int"))
pco2_product_biome_annual_anomaly_super_regions %>%
filter(year == 2023) %>%
mutate(
resid = paste(ifelse(
resid > 0, paste0("+", round(resid, 2)), round(resid, 2)
), round(resid_sd, 2), sep = "±"),
value = paste(ifelse(
value > 0, paste0("+", round(value, 2)), round(value, 2)
), round(value_sd, 2), sep = "±")
) %>%
select(-c(contains("_sd"), year)) %>%
pivot_wider(values_from = c(resid, value)) %>%
relocate(
biome,
value_temperature,
resid_temperature,
value_fgco2_int,
resid_fgco2_int,
value_fgco2,
resid_fgco2
) %>%
arrange(match(
biome,
c(
"NA-SPSS",
"NA-STPS",
"NA-STSS",
"North Atlantic",
"NP-SPSS",
"NP-STPS",
"NP-STSS",
"North Pacific",
"NH extratropics",
"PEQU-E",
"PEQU-W",
"AEQU",
"Equatorial Indian",
"Tropics",
"SA-STPS",
"SP-STPS",
"Southern Indian",
"SO-STSS",
"SH extratropics",
"Global non-polar",
"SO-SPSS",
"SO-ICE",
"Arctic",
"Global"
)
)) %>%
write_csv("../output/biome_anomaly_ensemble_mean_pco2_products.csv")pco2_product_biome_annual_anomaly_merged <-
full_join(region_biomes,
pco2_product_biome_annual_anomaly) %>%
mutate(region = case_when(biome == "Global non-polar" ~ "Global\nnon-polar",
region == "atlantic" ~ "Atlantic",
region == "pacific" ~ "Pacific",
region == "indian" ~ "Indian Ocean",
TRUE ~ region),
region = fct_rev(fct_inorder(region))) %>%
mutate(
latitude = case_when(
biome == "Global non-polar" ~ "Global\nnon-polar",
biome %in% c(
"NA-SPSS",
"NA-STSS",
"NA-STPS",
"NP-SPSS",
"NP-STSS",
"NP-STPS"
) ~ "NH extratropics",
biome %in% c(
"Equatorial Indian",
"PEQU-W",
"PEQU-E",
"AEQU"
) ~ "Tropics",
biome %in% c("SA-STPS", "SP-STPS", "Southern Indian", "SO-STSS") ~ "SH extratropics",
biome %in% c("SO-SPSS", "SO-ICE") ~ "SH polar",
biome %in% c("Arctic") ~ "NH polar",
TRUE ~ "other"
),
latitude = fct_relevel(latitude, c("Global\nnon-polar",
"NH polar",
"NH extratropics",
"Tropics",
"SH extratropics",
"SH polar"))) %>%
mutate(basin = case_when(
biome == "Global non-polar" ~ "",
str_detect(biome, "NA-|SA-|AEQU") ~ "Atlantic",
str_detect(biome, "NP-|SP-") ~ "Pacific",
str_detect(biome, "Indian") ~ "Indian",
str_detect(biome, "SO-") ~ "Southern\nOcean",
str_detect(biome, "Arctic") ~ "Arctic",
biome == "PEQU-E" ~ "Pacific-E",
biome == "PEQU-W" ~ "Pacific-W",
TRUE ~ "other")) %>%
mutate(biome_class = case_when(
str_detect(biome, "SPSS") ~ "Subpolar\nseasonally\nstratified\n(SPSS)",
str_detect(biome, "STSS") ~ "Subtropical\nseasonally\nstratified\n(STSS)",
str_detect(biome, "STPS|Southern Indian") ~ "Subtropical\npermanently\nstratified\n(STPS)",
str_detect(biome, "Arctic|ICE") ~ "Ice",
TRUE ~ ""),
biome_class = fct_relevel(biome_class,
"Subtropical\nseasonally\nstratified\n(STSS)",
after = 2)) %>%
filter(year == 2023,
name %in% c("temperature", "fgco2", "fgco2_int"))
pco2_product_biome_annual_anomaly_merged_ensemble <-
pco2_product_biome_annual_anomaly_merged %>%
filter(product %in% pco2_product_list) %>%
group_by(name, biome, basin, region, latitude, biome_class) %>%
summarise(resid_sd = sd(resid),
resid = mean(resid))
pco2_product_biome_annual_anomaly_merged_ensemble %>%
kable() %>%
kable_styling() %>%
scroll_box(height = "300px")| name | biome | basin | region | latitude | biome_class | resid_sd | resid | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fgco2 | AEQU | Atlantic | Atlantic | Tropics | 0.1277725 | -0.1073872 | |||||||||
| fgco2 | Arctic | Arctic | arctic | NH polar | Ice | 0.1218973 | 0.2860942 | ||||||||
| fgco2 | Equatorial Indian | Indian | Indian Ocean | Tropics | 0.0811651 | -0.0242599 | |||||||||
| fgco2 | Global non-polar | Global non-polar | |Global non-pola | seasonally stratified (SPSS) |
0.0820
permanently stratified (STP
|
) | 0.0306 | 92| 0.1359 | ||||||||
| fgco2 | NA-STSS | Atlantic | Atlantic | NH extratropics | Subtropical seasonally stratified (STSS |
0.1038
seasonally stratified (SPSS)
|
0.2998
permanently stratified (STP
|
) | 0.1258 | 40| 0.0990 | ||||||
| fgco2 | NP-STSS | Pacific | Pacific | NH extratropics | Subtropical seasonally stratified (STSS |
0.1121
permanently stratified (STP
|
) | 0.0616 | 08| -0.0603 | |||||||
| fgco2 | SO-ICE | Southern Ocean | |southern | |SH polar | |Ice |
0.197166
Ocean
|
|southern | |SH polar | |Subpolar seasonally stratified (SPSS) |
0.378
Ocean
|
|southern | |SH extratropics | |Subtropical seasonally stratified (STS | ) | 0.141 | 620| 0.017 |
| fgco2 | SP-STPS | Pacific | Pacific | SH extratropics | Subtropical permanently stratified (STP | ) | 0.1410 | 10| 0.0566 | ||||||||
| fgco2 | Southern Indian | Indian | Indian Ocean | SH extratropics | Subtropical permanently stratified (STP | ) | 0.1052 | 16| 0.1269 | ||||||||
| fgco2_int | AEQU | Atlantic | Atlantic | Tropics | 0.0128727 | -0.0108902 | |||||||||
| fgco2_int | Arctic | Arctic | arctic | NH polar | Ice | 0.0183644 | 0.0328425 | ||||||||
| fgco2_int | Equatorial Indian | Indian | Indian Ocean | Tropics | 0.0262191 | -0.0076427 | |||||||||
| fgco2_int | Global non-polar | Global non-polar | |Global non-pola | seasonally stratified (SPSS) |
0.0085
permanently stratified (STP
|
) | 0.0086 | 91| 0.0366 | ||||||||
| fgco2_int | NA-STSS | Atlantic | Atlantic | NH extratropics | Subtropical seasonally stratified (STSS |
0.0076
seasonally stratified (SPSS)
|
0.0480
permanently stratified (STP
|
) | 0.0648 | 94| 0.0513 | ||||||
| fgco2_int | NP-STSS | Pacific | Pacific | NH extratropics | Subtropical seasonally stratified (STSS |
0.0104
permanently stratified (STP
|
) | 0.0142 | 88| -0.0140 | |||||||
| fgco2_int | SO-ICE | Southern Ocean | |southern | |SH polar | |Ice |
0.042917
Ocean
|
|southern | |SH polar | |Subpolar seasonally stratified (SPSS) |
0.140
Ocean
|
|southern | |SH extratropics | |Subtropical seasonally stratified (STS | ) | 0.049 | 971| 0.006 |
| fgco2_int | SP-STPS | Pacific | Pacific | SH extratropics | Subtropical permanently stratified (STP | ) | 0.0926 | 02| 0.0372 | ||||||||
| fgco2_int | Southern Indian | Indian | Indian Ocean | SH extratropics | Subtropical permanently stratified (STP | ) | 0.0213 | 09| 0.0261 | ||||||||
| temperature | AEQU | Atlantic | Atlantic | Tropics | 0.0757248 | 0.2335304 | |||||||||
| temperature | Arctic | Arctic | arctic | NH polar | Ice | 0.0856725 | -0.0715219 | ||||||||
| temperature | Equatorial Indian | Indian | Indian Ocean | Tropics | 0.0572257 | 0.0140388 | |||||||||
| temperature | Global non-polar | Global non-polar | |Global non-pola | seasonally stratified (SPSS) |
0.0468
permanently stratified (STP
|
) | 0.0451 | 47| 0.4822 | ||||||||
| temperature | NA-STSS | Atlantic | Atlantic | NH extratropics | Subtropical seasonally stratified (STSS |
0.0287
seasonally stratified (SPSS)
|
0.0349
permanently stratified (STP
|
) | 0.0280 | 10| -0.0030 | ||||||
| temperature | NP-STSS | Pacific | Pacific | NH extratropics | Subtropical seasonally stratified (STSS |
0.0549
permanently stratified (STP
|
) | 0.0380 | 08| 0.1071 | |||||||
| temperature | SO-ICE | Southern Ocean | |southern | |SH polar | |Ice |
0.038917
Ocean
|
|southern | |SH polar | |Subpolar seasonally stratified (SPSS) |
0.035
Ocean
|
|southern | |SH extratropics | |Subtropical seasonally stratified (STS | ) | 0.065 | 036| 0.275 |
| temperature | SP-STPS | Pacific | Pacific | SH extratropics | Subtropical permanently stratified (STP | ) | 0.0281 | 54| 0.1133 | ||||||||
| temperature | Southern Indian | Indian | Indian Ocean | SH extratropics | Subtropical permanently stratified (STP | ) | 0.0905 | 71| 0.1065 |
pco2_product_biome_annual_anomaly_merged_ensemble %>%
filter(name != "fgco2_int", !str_detect(biome, "SO-SPSS|SO-ICE|Arctic")) %>%
ggplot(aes(x = basin, y = resid)) +
geom_hline(yintercept = 0) +
geom_col(aes(fill = "fCO2 product\nensemble mean"), col = "grey20") +
geom_linerange(aes(
ymin = resid - resid_sd,
ymax = resid + resid_sd,
col = "fCO2 product\nensemble SD"
)) +
scale_color_manual(values = "grey20", name = "") +
scale_fill_manual(values = "grey90", name = "") +
new_scale_color() +
geom_point(
data = pco2_product_biome_annual_anomaly_merged %>%
filter(
name != "fgco2_int",
product %in% pco2_product_list,
!str_detect(biome, "SO-SPSS|SO-ICE|Arctic")
),
aes(col = product, shape = product)
) +
scale_color_manual(values = color_products, name = "fCO2 products") +
scale_shape_manual(values = shape_products, name = "fCO2 products") +
new_scale_color() +
new_scale("shape") +
geom_point(
data = pco2_product_biome_annual_anomaly_merged %>%
filter(
name != "fgco2_int",
product %in% gobm_product_list,
!str_detect(biome, "SO-SPSS|SO-ICE|Arctic")
),
aes(col = product, shape = product),
position = position_nudge(x = 0.2)
) +
scale_color_manual(values = color_products, name = "GOBMs") +
scale_shape_manual(values = shape_products, name = "GOBMs") +
facet_nested(
name ~ latitude + biome_class,
scales = "free",
space = "free_x",
labeller = labeller(name = x_axis_labels),
switch = "y",
nest_line = element_line(linewidth = 0.8),
solo_line = TRUE,
strip = strip_nested(
text_x = list(
element_text(face = "bold"),
element_text(face = "bold"),
element_text(face = "bold"),
element_text(face = "bold"),
elem_list_text(),
elem_list_text(),
elem_list_text(),
elem_list_text(),
elem_list_text(),
elem_list_text(),
elem_list_text()
)
)
) +
theme(
axis.text.x = element_text(
angle = 90,
vjust = 0.5,
hjust = 1
),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
strip.background.x = element_blank()
)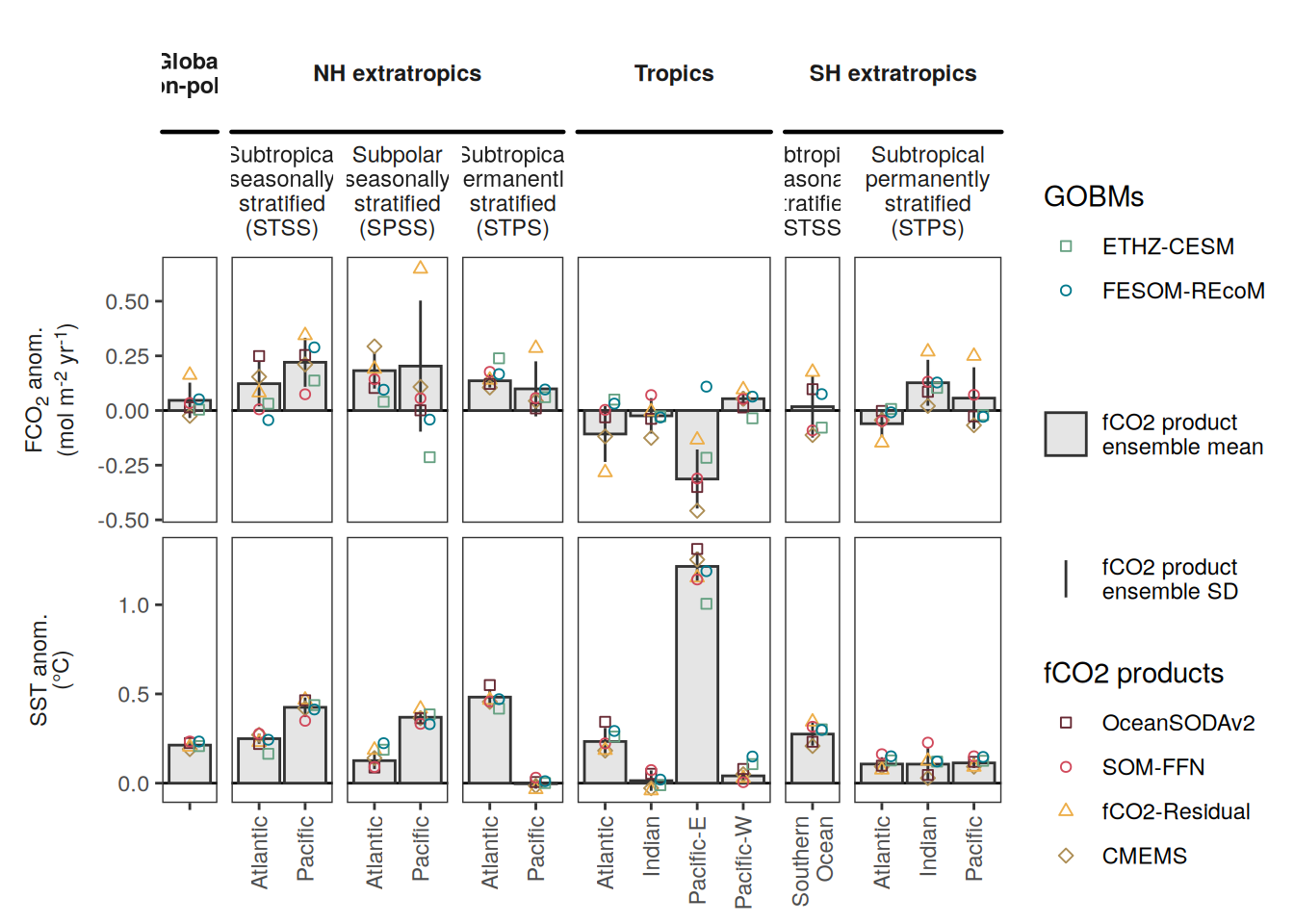
| Version | Author | Date |
|---|---|---|
| dd25c0c | jens-daniel-mueller | 2025-03-06 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
ggsave(width = 10,
height = 6,
dpi = 600,
filename = "../output/biome_anomaly_ensemble_mean_pco2_products.jpg")
p_global <- pco2_product_biome_annual_anomaly_merged_ensemble %>%
filter(biome == "Global non-polar") %>%
ggplot(aes(basin, resid)) +
geom_hline(yintercept = 0) +
geom_col(aes(fill = "fCO2 product\nensemble mean"), col = "grey20") +
geom_linerange(aes(ymin = resid - resid_sd,
ymax = resid + resid_sd,
col = "fCO2 product\nensemble SD")) +
scale_color_manual(values = "grey20", name = "") +
scale_fill_manual(values = "grey90", name = "") +
new_scale_color()+
geom_point(
data = pco2_product_biome_annual_anomaly_merged %>%
filter(biome == "Global non-polar",
product %in% pco2_product_list),
aes(col = product),
# position = position_nudge(x = -0.15),
shape = 21
) +
scale_color_manual(values = color_products,
name = "fCO2 products") +
new_scale_color()+
geom_point(data = pco2_product_biome_annual_anomaly_merged %>%
filter(biome == "Global",
product %in% gobm_product_list),
aes(col = product),
position = position_nudge(x = 0.2),
shape = 21) +
scale_color_manual(values = color_products,
name = "GOBMs") +
facet_nested(name ~ latitude + biome_class,
scales = "free", space = "free_x",
labeller = labeller(name = x_axis_labels),
switch = "y",
nest_line = element_line(),
solo_line = TRUE) +
theme(
axis.text.x = element_text(
angle = 90,
vjust = 0.5,
hjust = 1
),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
strip.background.x = element_blank(),
legend.position = "none"
)
p_biome <- pco2_product_biome_annual_anomaly_merged_ensemble %>%
filter(biome != "Global non-polar") %>%
ggplot(aes(basin, resid)) +
geom_hline(yintercept = 0) +
geom_col(aes(fill = "fCO2 product\nensemble mean"), col = "grey20") +
geom_linerange(aes(ymin = resid - resid_sd,
ymax = resid + resid_sd,
col = "fCO2 product\nensemble SD")) +
scale_color_manual(values = "grey20", name = "") +
scale_fill_manual(values = "grey90", name = "") +
new_scale_color()+
geom_point(
data = pco2_product_biome_annual_anomaly_merged %>%
filter(biome != "Global non-polar",
product %in% pco2_product_list),
aes(col = product),
# position = position_nudge(x = -0.15),
shape = 21
) +
scale_color_manual(values = color_products,
name = "fCO2 products") +
new_scale_color()+
geom_point(data = pco2_product_biome_annual_anomaly_merged %>%
filter(biome != "Global non-polar",
product %in% gobm_product_list),
aes(col = product),
position = position_nudge(x = 0.2),
shape = 21) +
scale_color_manual(values = color_products,
name = "GOBMs") +
facet_nested(name ~ latitude + biome_class,
scales = "free", space = "free_x",
labeller = labeller(name = ""),
# switch = "y",
nest_line = element_line(),
solo_line = TRUE
) +
theme(
axis.text.x = element_text(
angle = 90,
vjust = 0.5,
hjust = 1
),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
strip.text.y.right = element_text(colour = "transparent",
size = 0),
strip.placement = "outside",
strip.background.y = element_blank(),
strip.background.x = element_blank(),
legend.position = "bottom",
legend.direction = "vertical"
)
ggsave(cowplot::plot_grid(p_global, p_biome,
align = "hv",
axis = "tb",
rel_widths = c(1,7)),
width = 12,
height = 8,
dpi = 600,
filename = "../output/biome_anomaly_ensemble_mean_pco2_products_with_integrated_flux_and_SO.jpg")Seasonal anomalies
Annual mean trends
pco2_product_biome_monthly_detrended %>%
filter(biome %in% "NA-STPS",
name %in% c("temperature"),
product %in% pco2_product_list) %>%
select(-c(time, product)) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup()# A tibble: 408 × 7
year month biome name value fit resid
<dbl> <dbl> <chr> <fct> <dbl> <dbl> <dbl>
1 1990 1 NA-STPS temperature 23.3 23.4 -0.0403
2 1990 2 NA-STPS temperature 22.9 22.9 0.0672
3 1990 3 NA-STPS temperature 22.7 22.8 -0.0665
4 1990 4 NA-STPS temperature 23.2 23.2 -0.0419
5 1990 5 NA-STPS temperature 24.3 24.2 0.111
6 1990 6 NA-STPS temperature 25.3 25.3 -0.0603
7 1990 7 NA-STPS temperature 26.3 26.3 -0.0180
8 1990 8 NA-STPS temperature 27.0 27.0 0.0671
9 1990 9 NA-STPS temperature 27.3 27.2 0.131
10 1990 10 NA-STPS temperature 26.8 26.7 0.175
# ℹ 398 more rowspco2_product_biome_monthly_detrended %>%
filter(biome %in% "Global non-polar",
name %in% c("temperature"),
product %in% pco2_product_list) %>%
select(-c(time, product)) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup() %>%
# mutate(product = "Ensemble mean") %>%
p_season(title = NULL,
dim_col = NULL) +
theme(legend.position = "none")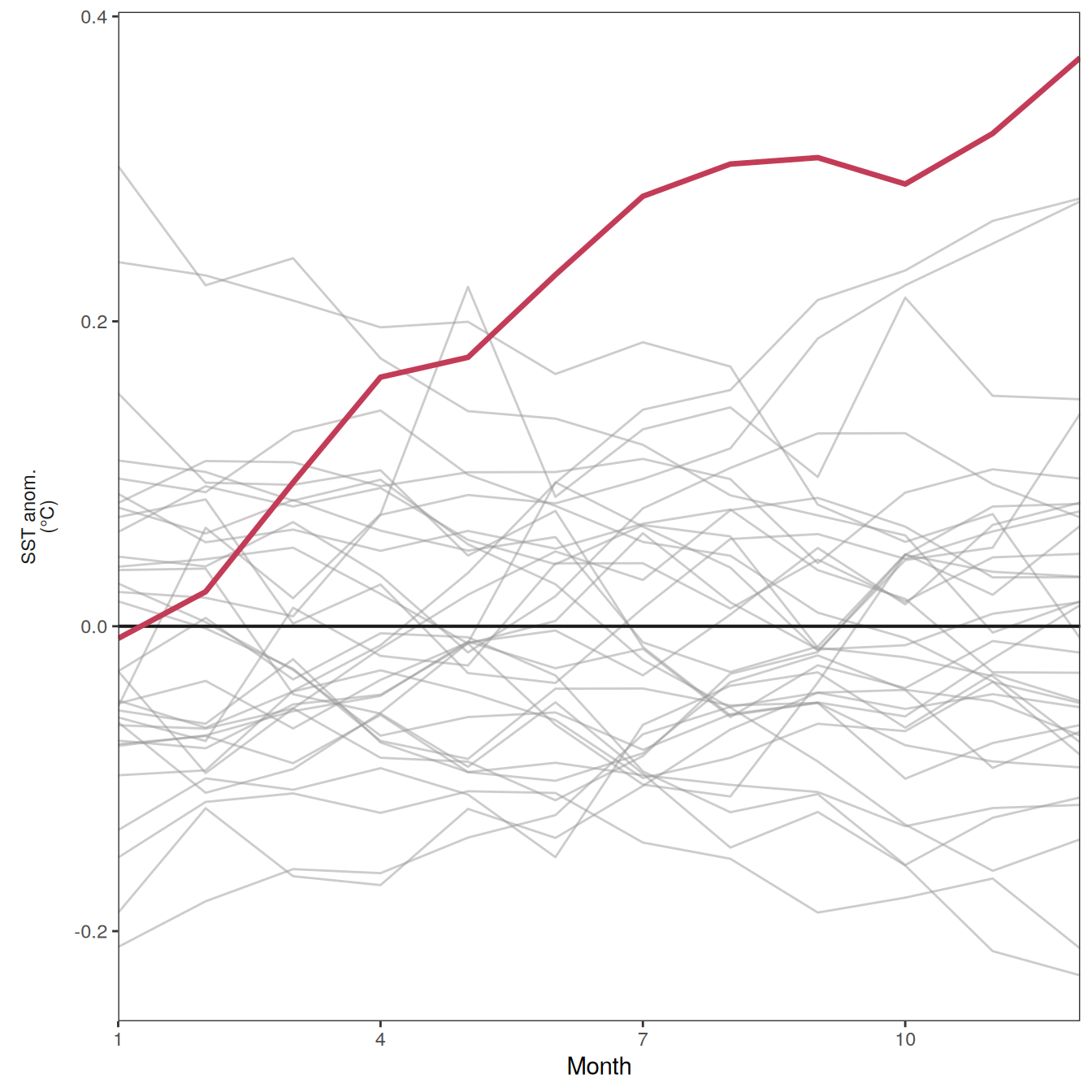
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
ggsave(width = 5,
height = 2,
dpi = 600,
filename = "../output/global_seasonality_sst_ensemble_mean_pco2_products.jpg")
pco2_product_biome_monthly_detrended %>%
filter(biome %in% "Global non-polar",
name %in% c("fgco2_int"),
product %in% pco2_product_list) %>%
select(-c(time, product)) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup() %>%
# mutate(product = "Ensemble mean") %>%
p_season(title = NULL,
dim_col = NULL) +
theme(legend.position = "none")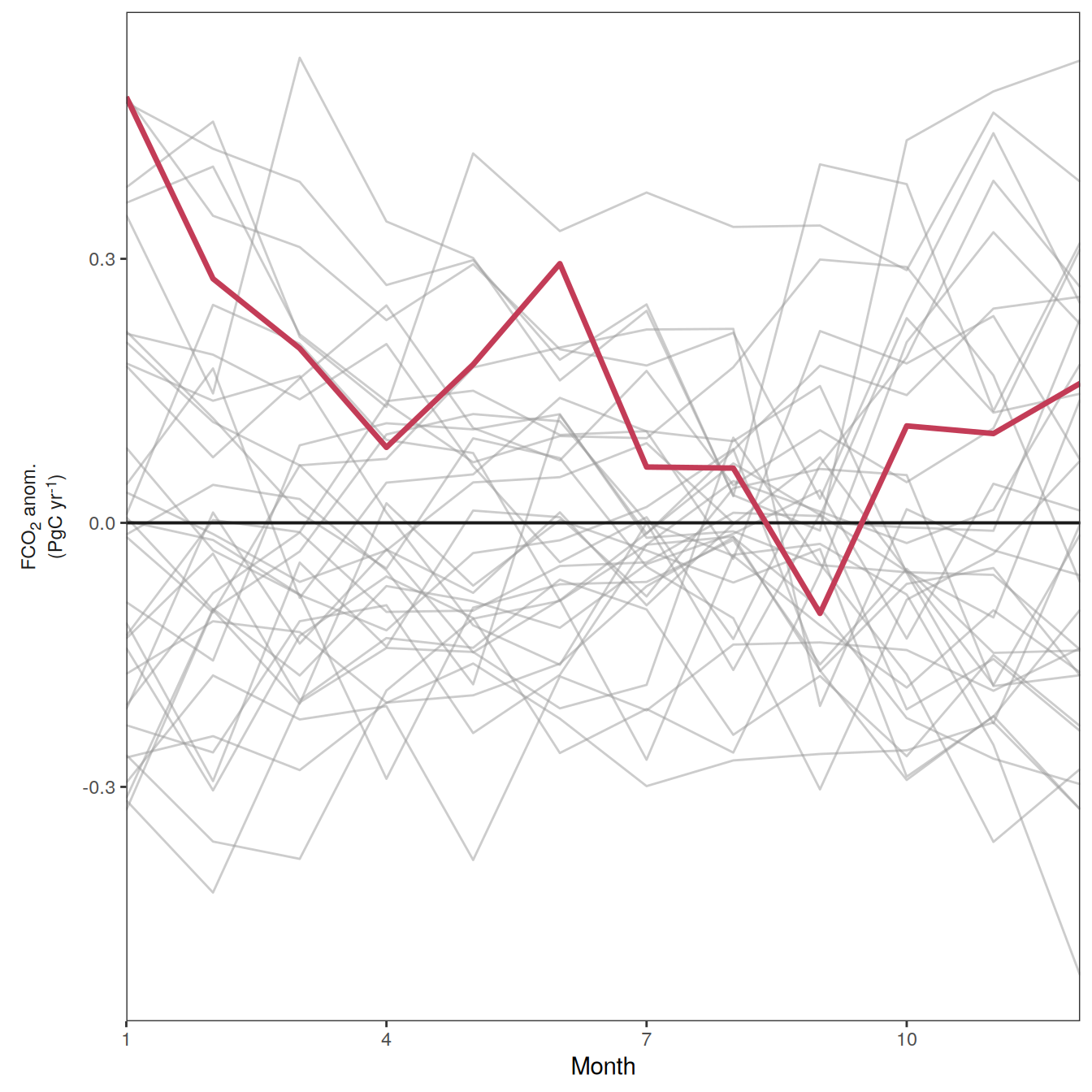
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
ggsave(width = 5,
height = 2,
dpi = 600,
filename = "../output/global_seasonality_fgco2_ensemble_mean_pco2_products.jpg")pco2_product_biome_annual_detrended %>%
filter(biome %in% "Global non-polar",
name %in% c("fgco2", "temperature")) %>%
p_season(title = NULL) +
theme(legend.position = "top")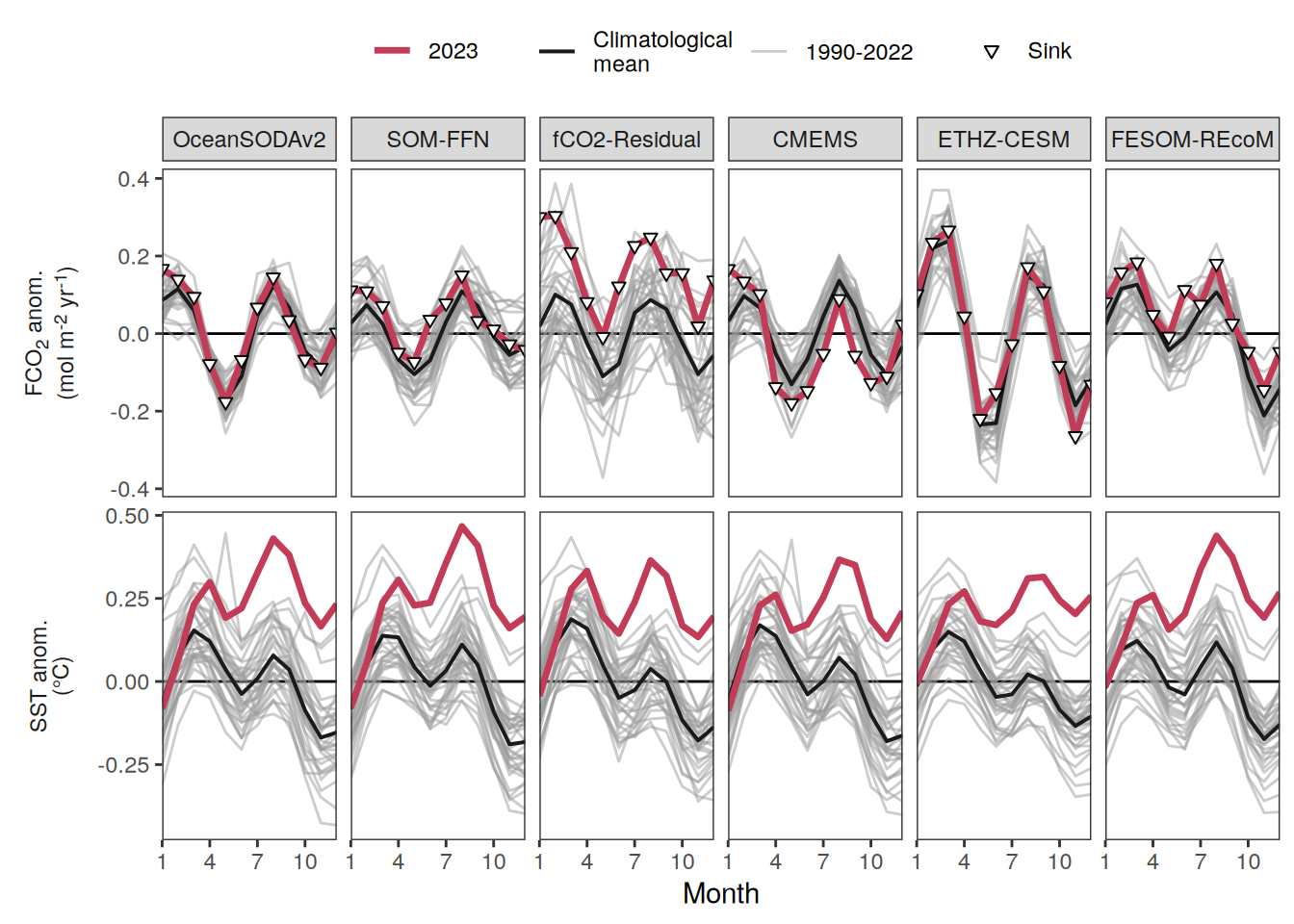
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7919445 | jens-daniel-mueller | 2024-05-31 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| 7c448f7 | jens-daniel-mueller | 2024-05-31 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 1eefab2 | jens-daniel-mueller | 2024-05-21 |
ggsave(width = 10,
height = 4,
dpi = 600,
filename = "../output/global_seasonality_sst_all_products.jpg")pco2_product_biome_annual_detrended %>%
filter(biome %in% "Global non-polar", name %in% name_core) %>%
p_season(title = "Anomalies from predicted annual mean | Global non-polar")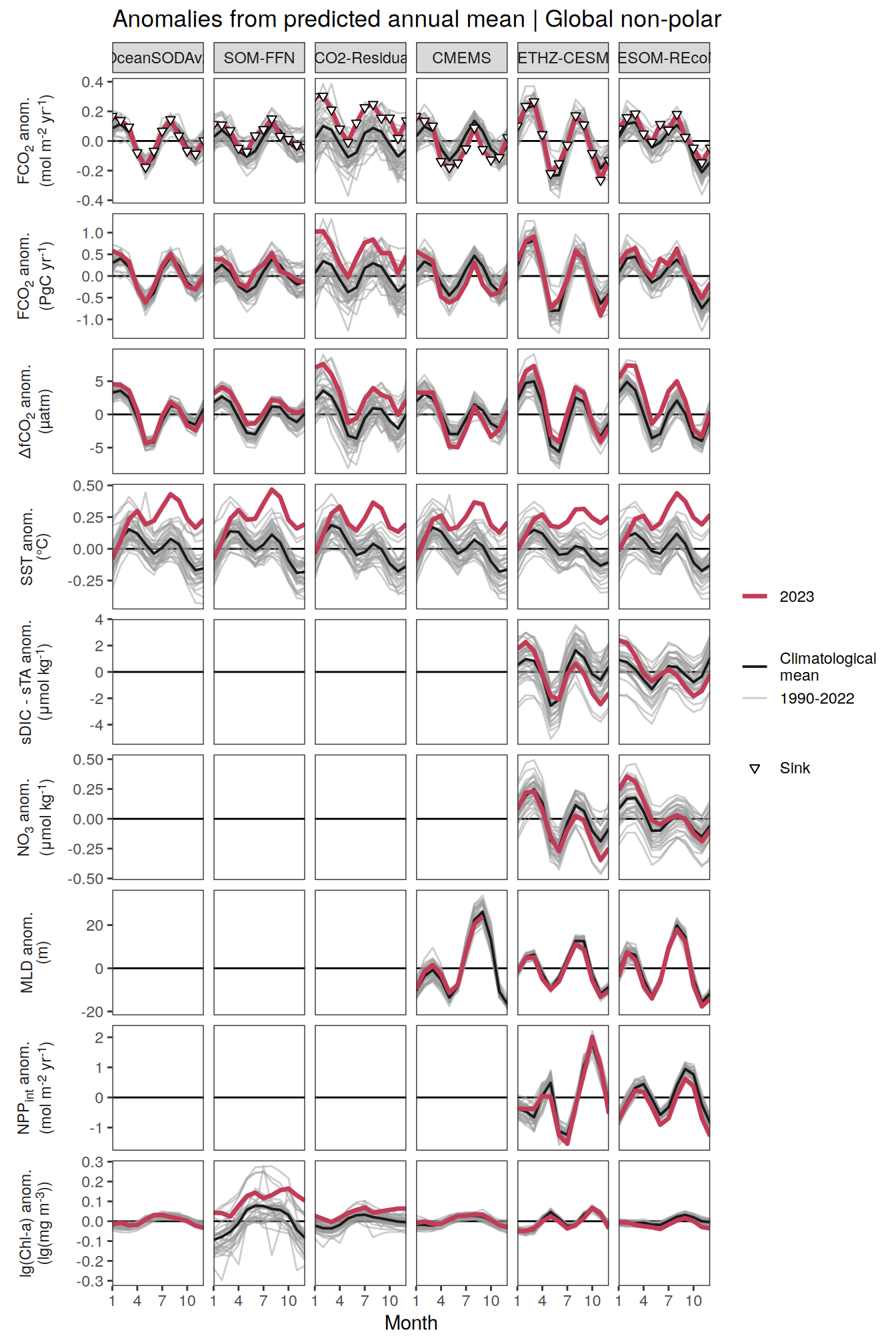
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2a28b07 | jens-daniel-mueller | 2024-07-22 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7919445 | jens-daniel-mueller | 2024-05-31 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 7f9c687 | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| 238d229 | jens-daniel-mueller | 2024-04-11 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
pco2_product_biome_annual_detrended %>%
filter(biome %in% key_biomes,
name %in% name_core) %>%
group_split(biome) %>%
# head(1) %>%
map(~ p_season(
df = .x,
title = paste("Anomalies from predicted annual mean |", .x$biome)
))[[1]]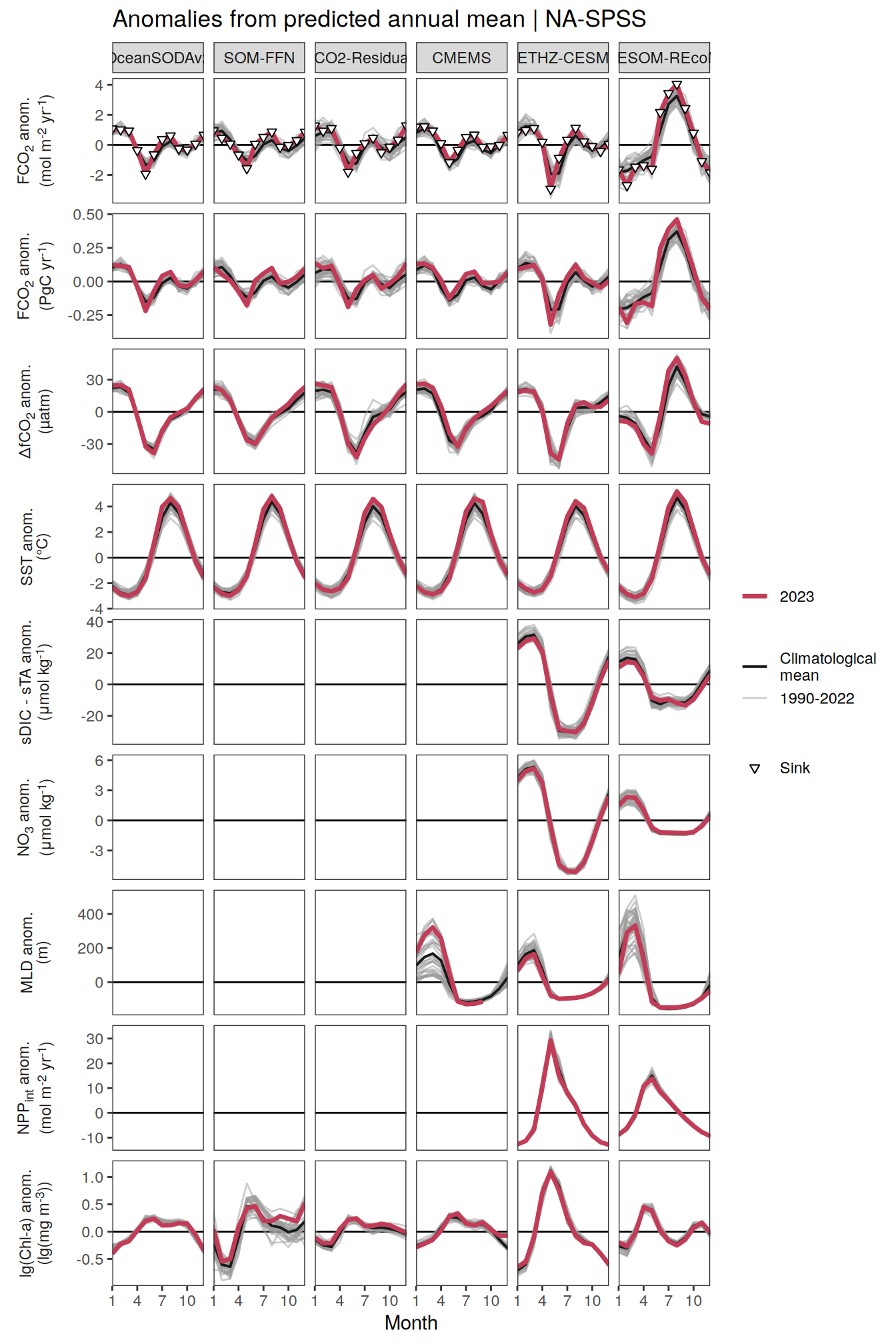
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 2a28b07 | jens-daniel-mueller | 2024-07-22 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| e44a62b | jens-daniel-mueller | 2024-04-23 |
| 7f9c687 | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| 238d229 | jens-daniel-mueller | 2024-04-11 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
[[2]]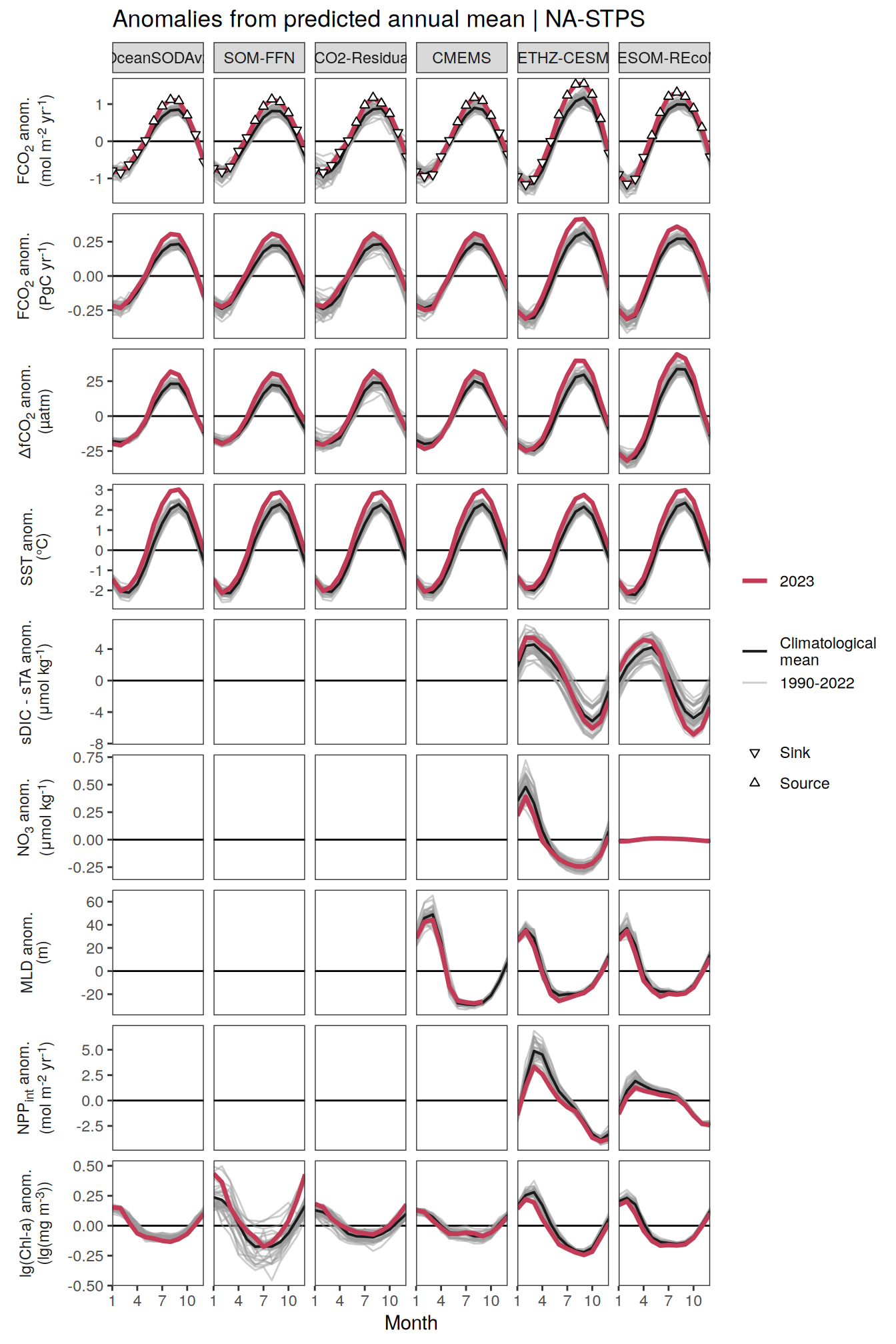
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 2a28b07 | jens-daniel-mueller | 2024-07-22 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7919445 | jens-daniel-mueller | 2024-05-31 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| 238d229 | jens-daniel-mueller | 2024-04-11 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
[[3]]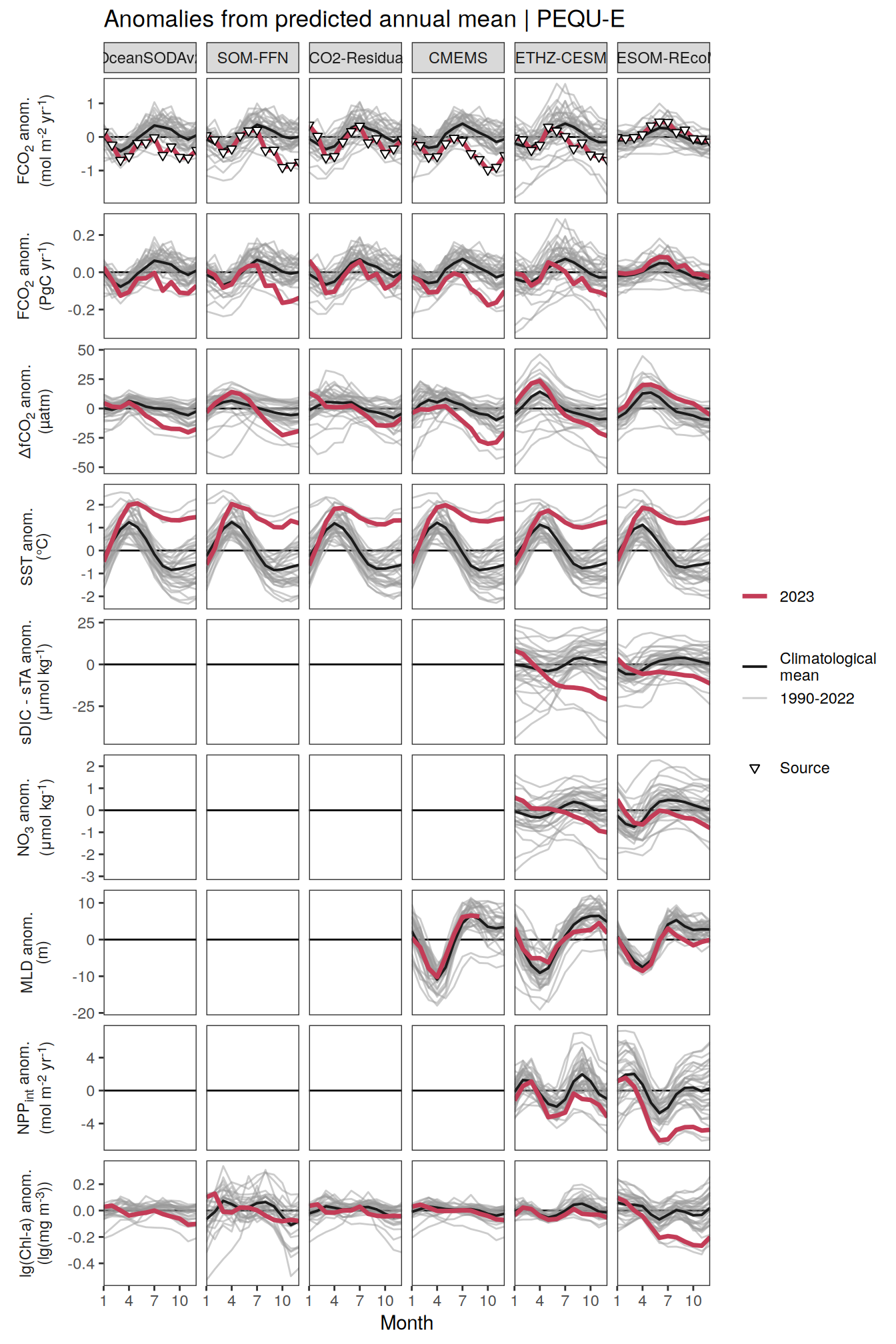
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 2a28b07 | jens-daniel-mueller | 2024-07-22 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 7919445 | jens-daniel-mueller | 2024-05-31 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| 238d229 | jens-daniel-mueller | 2024-04-11 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
Monthly mean trends
grDevices::cairo_pdf("../output/biome_seasonality_all_products.pdf",
width = 9,
height = 10,
onefile = TRUE)
pco2_product_biome_monthly_detrended %>%
filter(biome %in% c("Global non-polar", key_biomes),
name %in% name_core) %>%
group_split(biome) %>%
# head(1) %>%
map(~ p_season(
df = .x,
title = paste("Anomalies from predicted monthly mean |", .x$biome)
))[[1]]
[[2]]
[[3]]
[[4]]dev.off()png
2 Flux anomaly correlation
The following plots aim to unravel the correlation between biome-, super-biome- or globally- integrated monthly flux anomalies and the corresponding anomalies of the means/integrals of each other variable.
Anomalies are first presented are first presented in absolute units. Due to the different flux magnitudes, we need to plot the globally and biome-integrated fluxes separately. Secondly, we normalize the anomalies to the monthly spread (expressed as standard deviation) of the anomalies from 1990 to 2021.
Annual anomalies
Absolute
pco2_product_biome_annual_anomaly %>%
filter(biome %in% c("Global non-polar", key_biomes),
name %in% name_core) %>%
mutate(biome = if_else(biome == "Global non-polar", "Global non-polar", biome)) %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(product, year, biome, fgco2_int)) %>%
filter(name == "temperature") %>%
group_split(name) %>%
# tail(1) %>%
map(
~ ggplot(data = .x,
aes(value, fgco2_int)) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_point(
data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(fill = "1990-2022"),
shape = 21
) +
scale_color_manual(values = "grey60", name = "X") +
scale_fill_manual(values = "grey60", name = "X") +
new_scale_fill() +
new_scale_color() +
geom_point(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(fill = as.factor(year)),
shape = 21,
size = 3
) +
scale_fill_manual(
values = rev(warm_cool_gradient[c(17,13,20)]),
guide = guide_legend(reverse = TRUE,
order = 2)
) +
scale_color_manual(
values = rev(warm_cool_gradient[c(17,13,20)]),
guide = guide_legend(reverse = TRUE,
order = 2)
) +
labs(y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks(unique(.x$name))$i_legend_title) +
facet_grid2(
product ~ biome,
scales = "free",
independent = "y"
) +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)
)[[1]]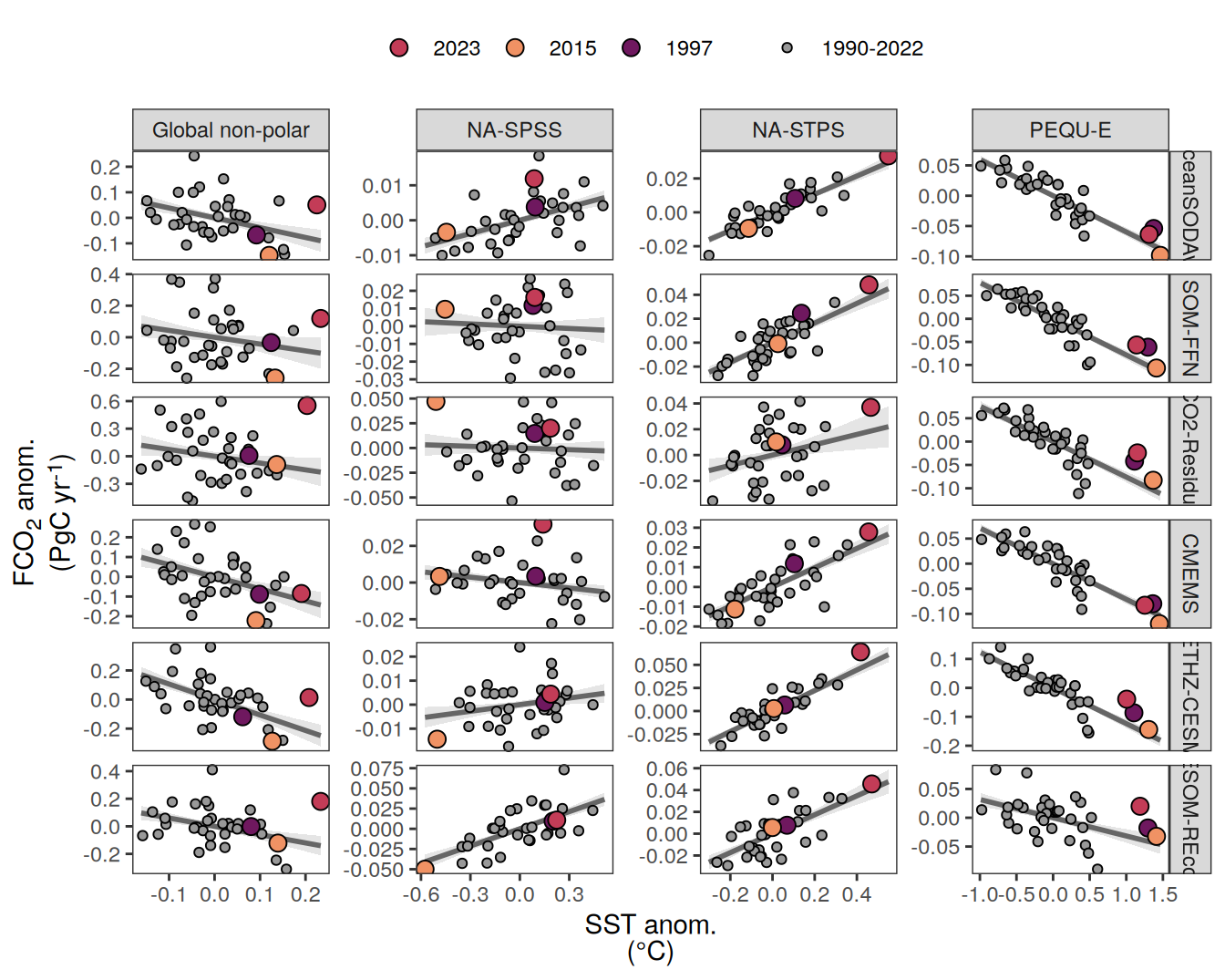
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 0493049 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
ggsave(width = 8,
height = 10,
dpi = 600,
filename = "../output/biome_anomaly_correlation_all_pco2_products.jpg")
pco2_product_biome_annual_anomaly_ensemble <-
pco2_product_biome_annual_anomaly %>%
filter(name %in% name_core, product %in% pco2_product_list) %>%
select(-c(value, fit, product)) %>%
fgroup_by(name, biome, year) %>%
fsummarise(sd = fsd(resid),
mean = fmean(resid))
pco2_product_biome_annual_anomaly_ensemble <-
full_join(
pco2_product_biome_annual_anomaly_ensemble %>%
filter(name == "fgco2_int") %>%
pivot_wider(values_from = c(sd, mean)),
pco2_product_biome_annual_anomaly_ensemble %>%
filter(name != "fgco2_int")
)
pco2_product_biome_annual_anomaly_super_regions %>%
filter(name %in% c("fgco2_int", "temperature")) %>%
select(-contains("value")) %>%
pivot_wider(values_from = contains("resid")) %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
ggplot(aes(resid_temperature, resid_fgco2_int)) +
# geom_vline(xintercept = 0) +
# geom_hline(yintercept = 0) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
)+
geom_linerange(
data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(
ymin = resid_fgco2_int - resid_sd_fgco2_int,
ymax = resid_fgco2_int + resid_sd_fgco2_int,
col = "1990-2022"
)
) +
geom_linerange(
data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(
xmin = resid_temperature - resid_sd_temperature,
xmax = resid_temperature + resid_sd_temperature,
col = "1990-2022"
)
) +
geom_point(data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(fill = "1990-2022"),
shape = 21) +
scale_color_manual(values = "grey60", name = "X") +
scale_fill_manual(values = "grey60", name = "X") +
new_scale_fill() +
new_scale_color() +
geom_linerange(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(
ymin = resid_fgco2_int - resid_sd_fgco2_int,
ymax = resid_fgco2_int + resid_sd_fgco2_int,
col = as.factor(year)
),
linewidth = 1
) +
geom_linerange(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(
xmin = resid_temperature - resid_sd_temperature,
xmax = resid_temperature + resid_sd_temperature,
col = as.factor(year)
),
linewidth = 1
) +
geom_point(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(fill = as.factor(year)),
shape = 21,
size = 3
) +
scale_fill_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
scale_color_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
labs(y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks(unique("temperature"))$i_legend_title) +
facet_wrap(~ biome, scales = "free") +
# theme_classic() +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank()
# strip.background = element_blank()
)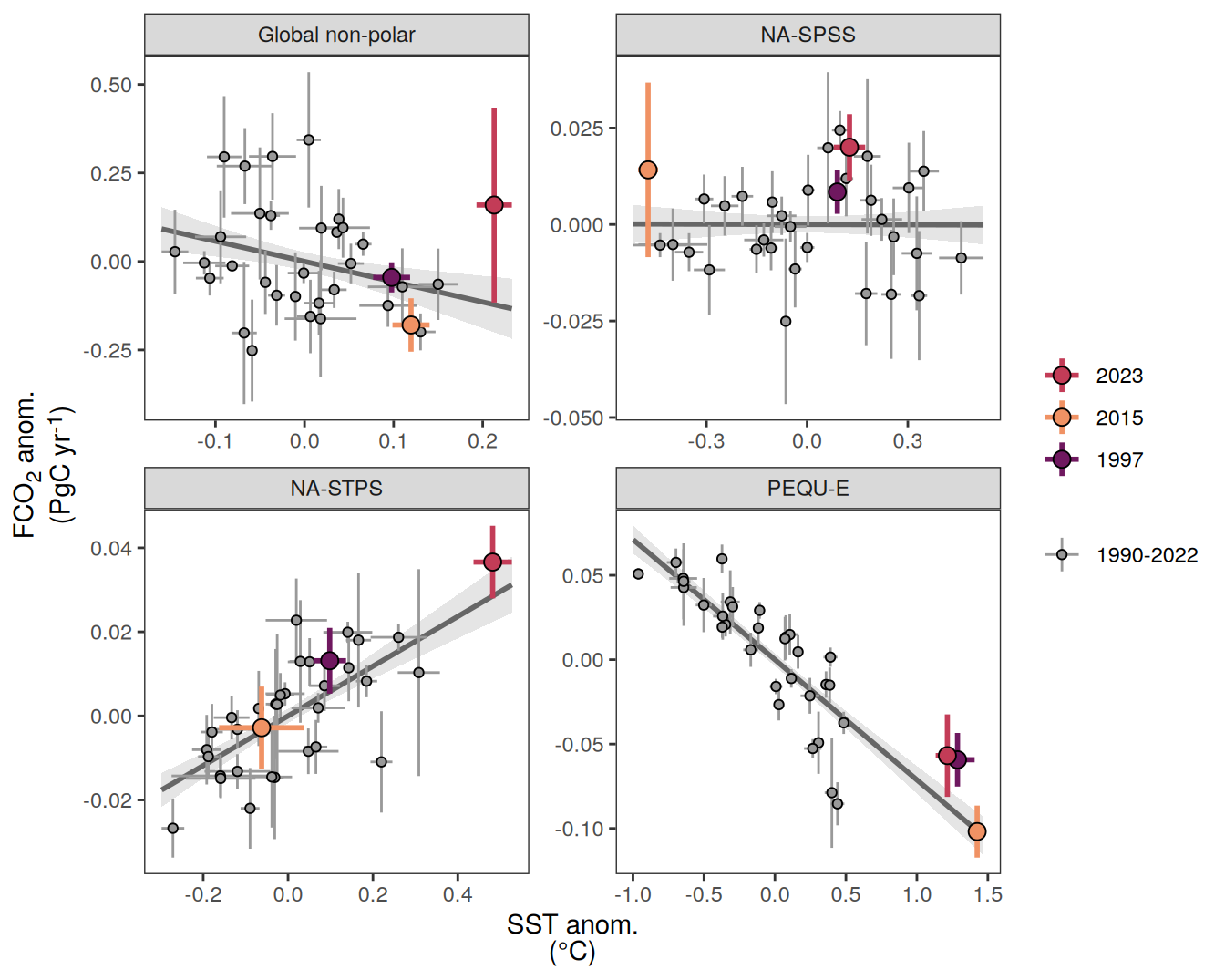
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| dd97823 | jens-daniel-mueller | 2024-06-28 |
| c6f967e | jens-daniel-mueller | 2024-06-28 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| 0493049 | jens-daniel-mueller | 2024-05-29 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
ggsave(width = 8,
height = 6,
dpi = 600,
filename = "../output/biome_anomaly_correlation_ensemble_mean_pco2_products.jpg")
pco2_product_biome_annual_anomaly_super_regions %>%
filter(name %in% c("fgco2_int", "temperature")) %>%
select(-contains("value")) %>%
pivot_wider(values_from = contains("resid")) %>%
ggplot(aes(resid_temperature, resid_fgco2_int)) +
# geom_vline(xintercept = 0) +
# geom_hline(yintercept = 0) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_linerange(
data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(
ymin = resid_fgco2_int - resid_sd_fgco2_int,
ymax = resid_fgco2_int + resid_sd_fgco2_int,
col = "1990-2022"
)
) +
geom_linerange(
data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(
xmin = resid_temperature - resid_sd_temperature,
xmax = resid_temperature + resid_sd_temperature,
col = "1990-2022"
)
) +
geom_point(data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(fill = "1990-2022"),
shape = 21) +
scale_color_manual(values = "grey60", name = "X") +
scale_fill_manual(values = "grey60", name = "X") +
new_scale_fill() +
new_scale_color() +
geom_linerange(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(
ymin = resid_fgco2_int - resid_sd_fgco2_int,
ymax = resid_fgco2_int + resid_sd_fgco2_int,
col = as.factor(year)
),
linewidth = 1
) +
geom_linerange(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(
xmin = resid_temperature - resid_sd_temperature,
xmax = resid_temperature + resid_sd_temperature,
col = as.factor(year)
),
linewidth = 1
) +
geom_point(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(fill = as.factor(year)),
shape = 21,
size = 3
) +
scale_fill_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
scale_color_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
labs(y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks(unique("temperature"))$i_legend_title) +
facet_wrap(~ biome, scales = "free",
ncol = 4) +
# theme_classic() +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)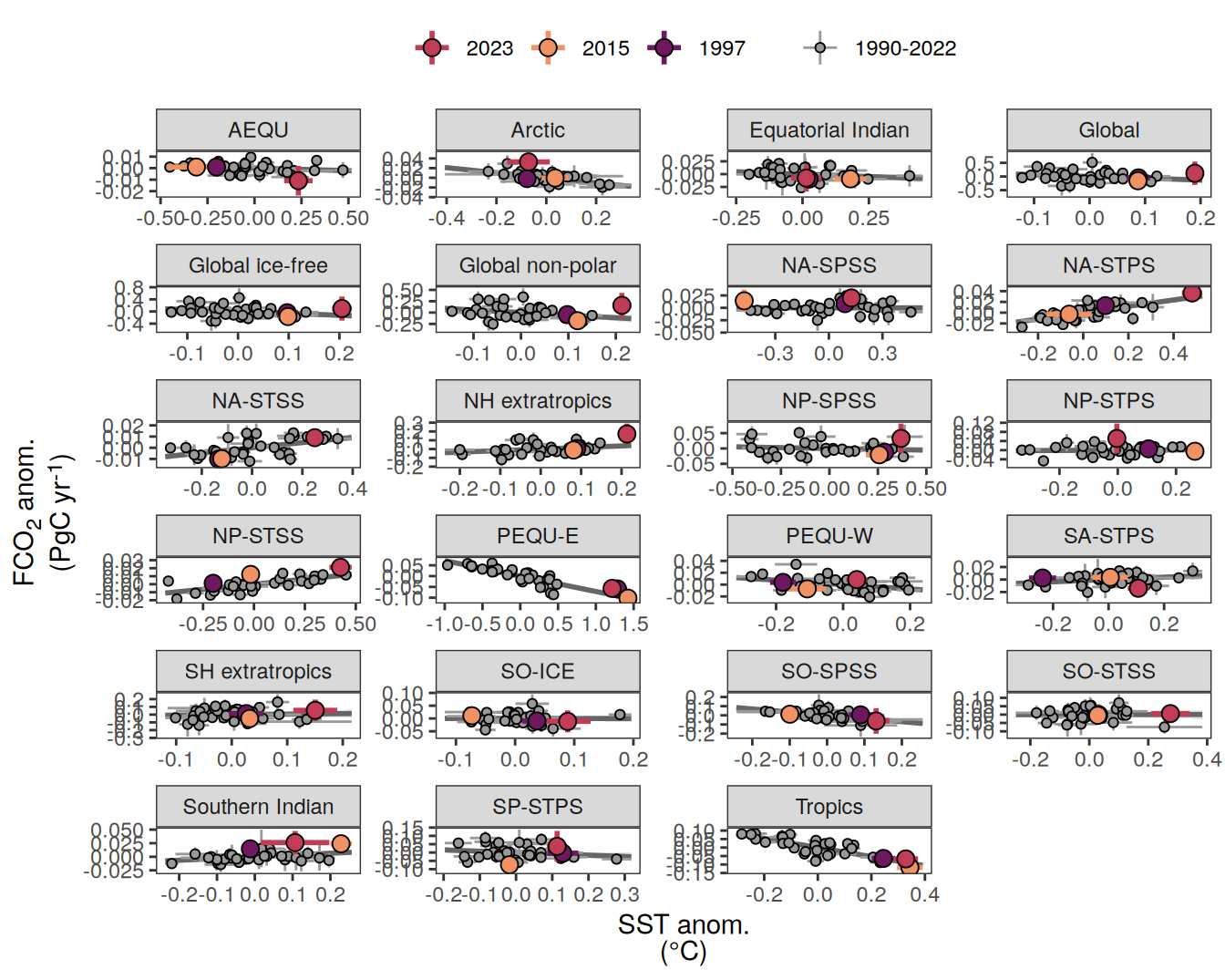
| Version | Author | Date |
|---|---|---|
| b9e8f8c | jens-daniel-mueller | 2025-03-10 |
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 4acb1fc | jens-daniel-mueller | 2024-09-05 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| 0493049 | jens-daniel-mueller | 2024-05-29 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
ggsave(width = 9,
height = 12,
dpi = 600,
filename = "../output/biome_anomaly_correlation_ensemble_mean_pco2_products_all_biomes.jpg")
pco2_product_biome_annual_anomaly %>%
filter(
biome %in% c("Global non-polar", key_biomes),
name %in% c(
"fgco2_int",
"chl",
"dfco2",
"sfco2",
"atm_fco2",
"temperature",
"sdissic",
"no3",
"int_pp",
"mld",
"kw_sol"
)
) %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(product, year, biome, fgco2_int)) %>%
group_by(product, name, biome) %>%
summarise(correlation = cor(fgco2_int, value)) %>%
ungroup() %>%
group_by(name) %>%
mutate(correlation_mean = mean(abs(correlation), na.rm = TRUE)) %>%
ungroup() %>%
mutate(name = fct_reorder(name, correlation_mean)) %>%
ggplot(aes(product,name,fill=correlation)) +
geom_tile() +
scale_fill_divergent() +
facet_wrap(~ biome) +
labs(title = "Correlation with FCO2 on a annual mean basis") +
theme(axis.text.x = element_text(angle = 90, hjust = 1),
axis.title = element_blank(),
legend.position = c(0.85,0.1),
legend.direction = "horizontal")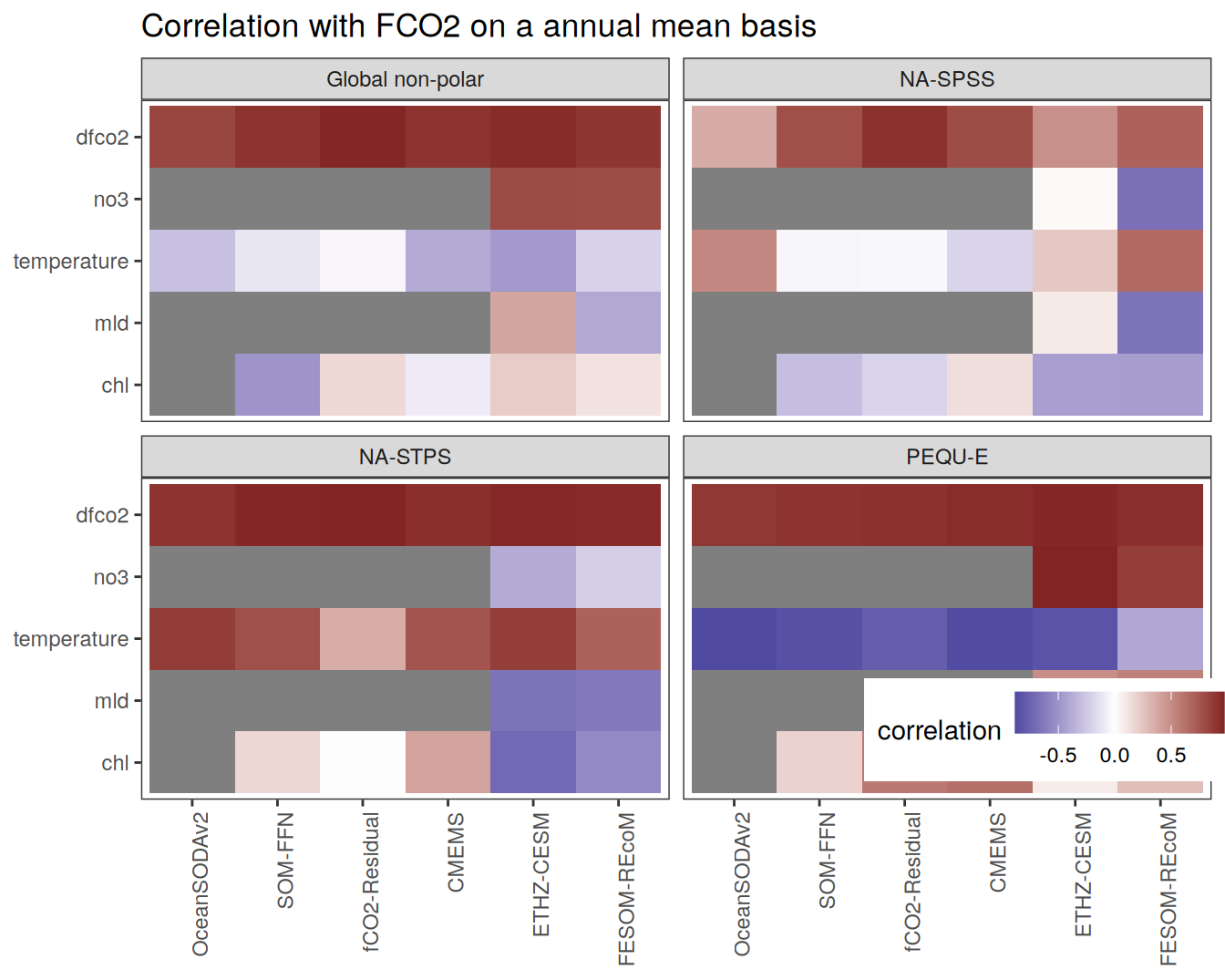
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| 0493049 | jens-daniel-mueller | 2024-05-29 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
pco2_product_biome_monthly_anomaly %>%
filter(
biome %in% c("Global non-polar", key_biomes),
name %in% c(
"fgco2_int",
"chl",
"dfco2",
"sfco2",
"atm_fco2",
"temperature",
"sdissic",
"no3",
"int_pp",
"mld",
"kw_sol"
)
) %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(product, year, month, biome, fgco2_int)) %>%
group_by(product, name, biome) %>%
summarise(correlation = cor(fgco2_int, value)) %>%
ungroup() %>%
group_by(name) %>%
mutate(correlation_mean = mean(abs(correlation), na.rm = TRUE)) %>%
ungroup() %>%
mutate(name = fct_reorder(name, correlation_mean)) %>%
ggplot(aes(product,name,fill=correlation)) +
geom_tile() +
scale_fill_divergent() +
facet_wrap(~ biome) +
labs(title = "Correlation with FCO2 on a monthly mean basis") +
theme(axis.text.x = element_text(angle = 90, hjust = 1),
axis.title = element_blank(),
legend.position = c(0.85,0.1),
legend.direction = "horizontal")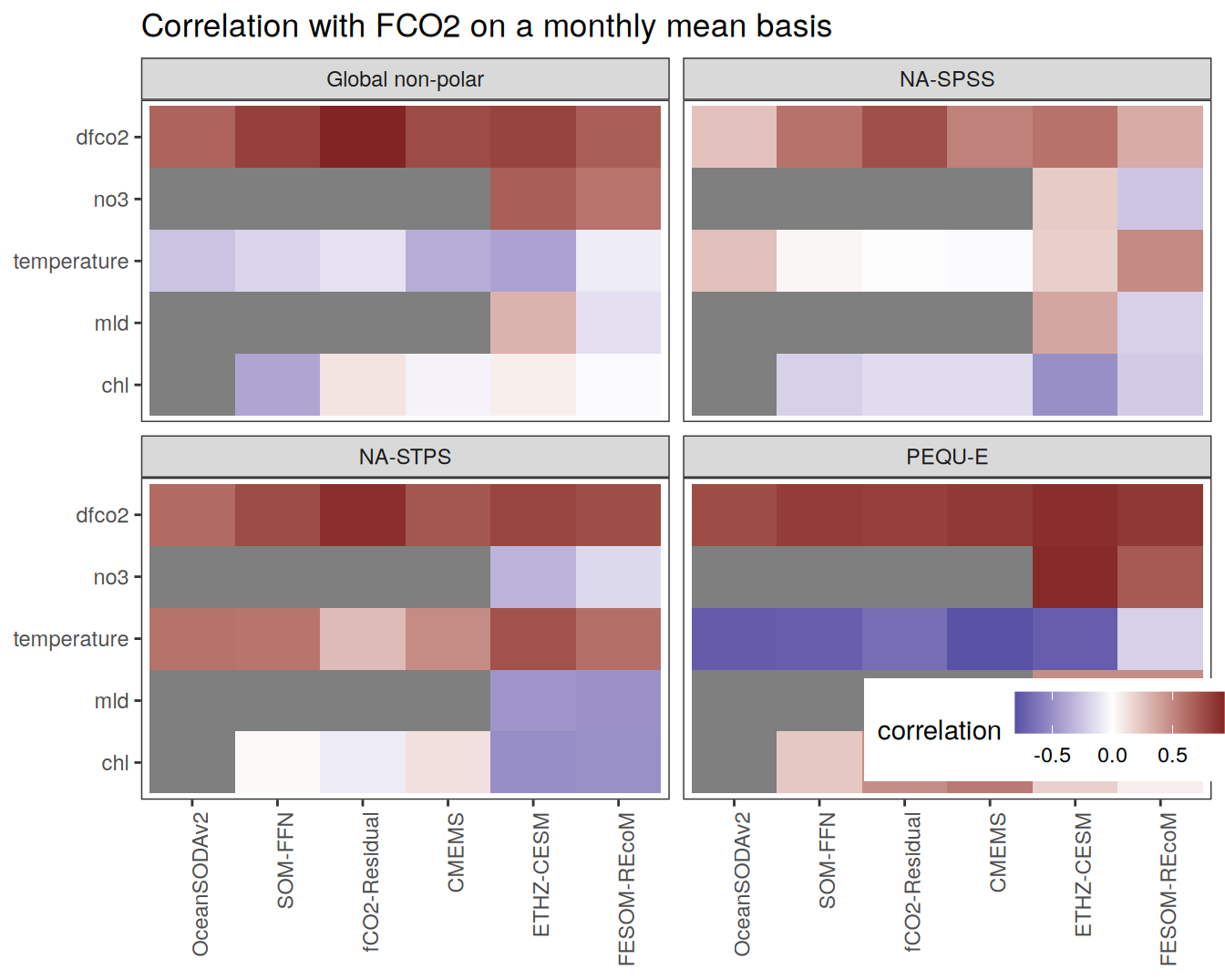
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 478e699 | jens-daniel-mueller | 2024-06-14 |
| 0493049 | jens-daniel-mueller | 2024-05-29 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
pco2_product_biome_monthly_anomaly %>%
filter(
biome %in% c("Global non-polar", key_biomes),
name %in% c(
"fgco2_int",
"chl",
"dfco2",
"sfco2",
"atm_fco2",
"temperature",
"sdissic",
"no3",
"int_pp",
"mld",
"kw_sol"
)
) %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(product, year, month, biome, fgco2_int)) %>%
group_by(product, name, biome, month) %>%
summarise(correlation = cor(fgco2_int, value)) %>%
ungroup() %>%
group_by(name) %>%
mutate(correlation_mean = mean(abs(correlation), na.rm = TRUE)) %>%
ungroup() %>%
mutate(name = fct_reorder(name, correlation_mean)) %>%
ggplot(aes(month, correlation, col = name)) +
geom_hline(yintercept = 0) +
geom_path() +
facet_grid(product ~ biome) +
labs(title = "Correlation with FCO2 on a monthly mean basis")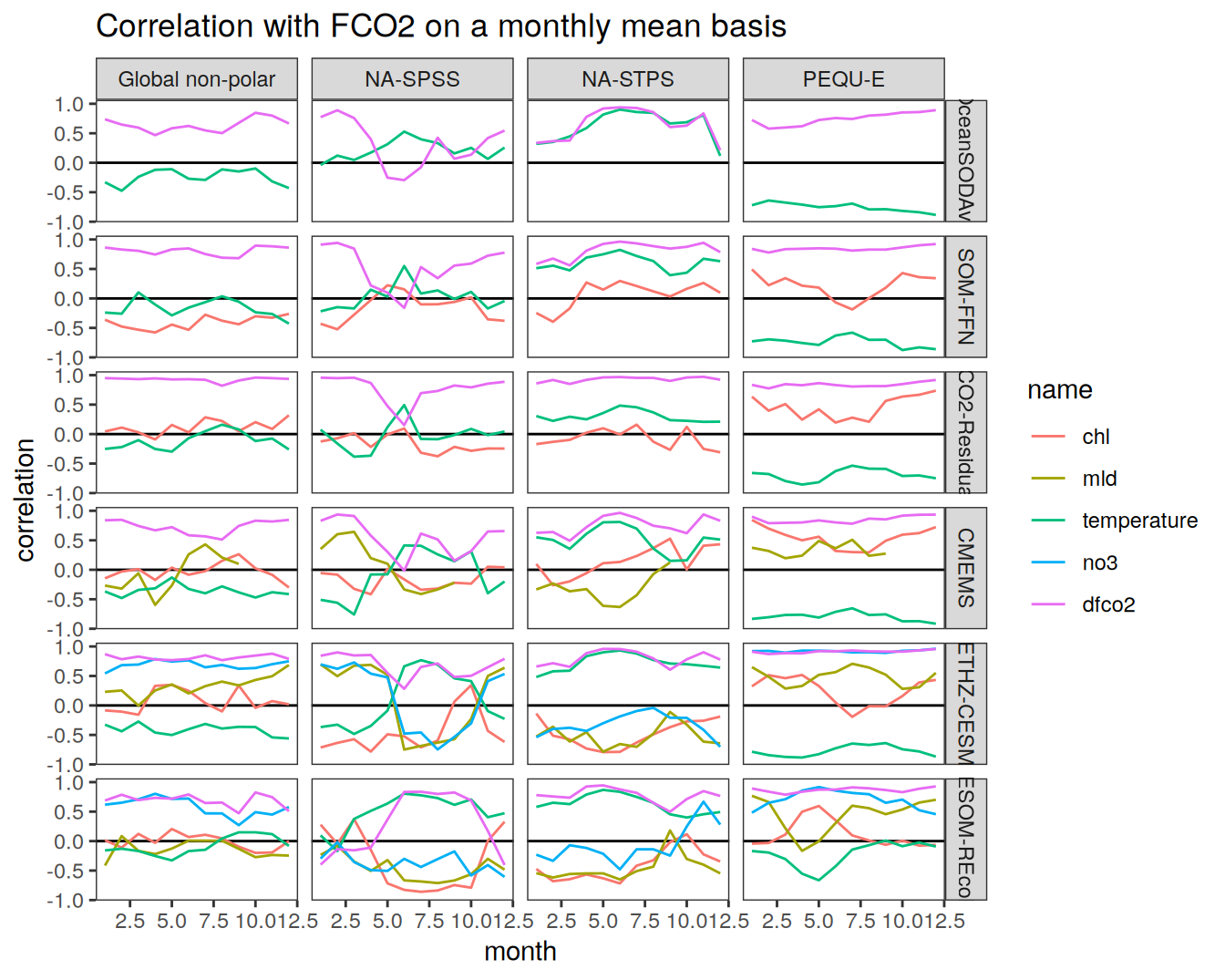
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 0493049 | jens-daniel-mueller | 2024-05-29 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 589243f | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 3fea035 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| dfcf790 | jens-daniel-mueller | 2024-04-11 |
| d5075c5 | jens-daniel-mueller | 2024-04-11 |
| 19f40c9 | jens-daniel-mueller | 2024-04-05 |
| a83c8fc | jens-daniel-mueller | 2024-04-03 |
SST based prediction
pco2_product_biome_annual_anomaly_temperature_predict <-
full_join(
pco2_product_biome_annual_anomaly_temperature_predict,
pco2_product_biome_annual_anomaly_temperature_predict %>%
filter(year != 2023) %>%
nest(data = -c(product, biome)) %>%
mutate(fit = map(
data, ~ flm(formula = fgco2_int ~ temperature, data = .x)
)) %>%
unnest_wider(fit) %>%
select(product, biome, slope = temperature) %>%
mutate(slope = as.vector(slope))
)
pco2_product_biome_annual_anomaly_temperature_predict <-
pco2_product_biome_annual_anomaly_temperature_predict %>%
mutate(fgco2_predict_int_biome = slope * temperature)
pco2_product_biome_annual_anomaly_temperature_predict %>%
select(product,
year,
biome,
`true anomaly` = fgco2_int,
`SST pattern` = fgco2_predict_int,
`SST mean` = fgco2_predict_int_biome) %>%
pivot_longer(4:6,
values_to = "resid") %>%
filter(biome %in% c("Global non-polar")) %>%
ggplot(aes(year, resid))+
geom_hline(yintercept = 0) +
geom_path(aes(col = name))+
geom_point(aes(fill = name), shape = 21, size = 1) +
labs(y = labels_breaks("fgco2_int")$i_legend_title) +
facet_wrap( ~ product) +
theme(
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
legend.title = element_blank()
)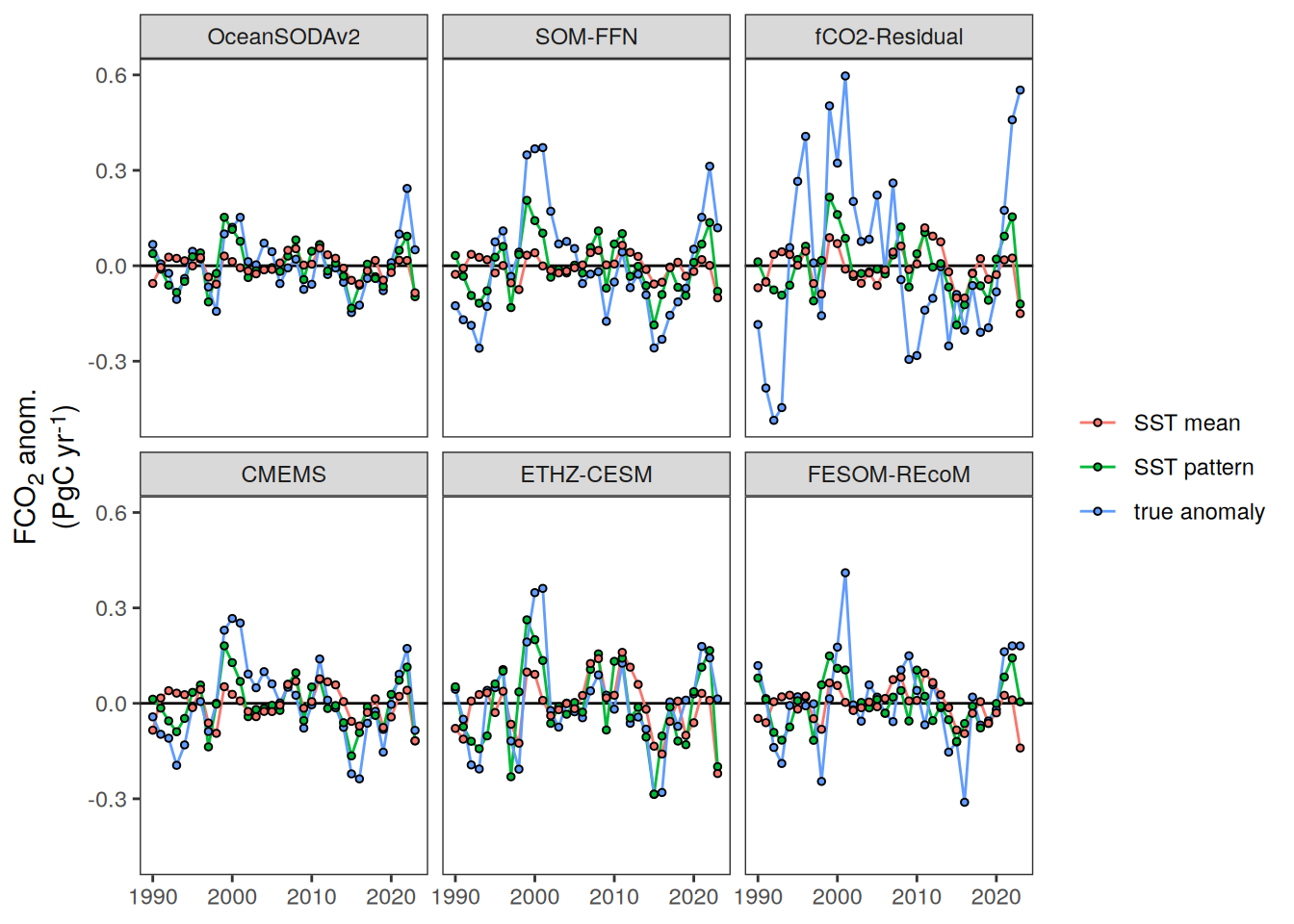
ggsave(width = 10,
height = 6,
dpi = 600,
filename = "../output/biome_flux_anomaly_prediction_timeseries_all_pco2_products_global.jpg")
pco2_product_biome_annual_anomaly_temperature_predict %>%
select(product,
year,
biome,
fgco2_int,
`SST pattern` = fgco2_predict_int,
`SST mean` = fgco2_predict_int_biome) %>%
pivot_longer(contains("SST"),
values_to = "resid") %>%
filter(biome %in% c("Global non-polar")) %>%
ggplot(aes(fgco2_int, resid))+
geom_vline(xintercept = 0) +
geom_hline(yintercept = 0) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_point(data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(fill = "1990-2022"),
shape = 21) +
scale_color_manual(values = "grey60", name = "X") +
scale_fill_manual(values = "grey60", name = "X") +
new_scale_fill() +
new_scale_color() +
geom_point(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(fill = as.factor(year)),
shape = 21,
size = 3
) +
scale_fill_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
scale_color_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
labs(x = labels_breaks("fgco2_int")$i_legend_title,
y = labels_breaks("fgco2_predict_int")$i_legend_title) +
facet_grid(name ~ product) +
coord_equal() +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)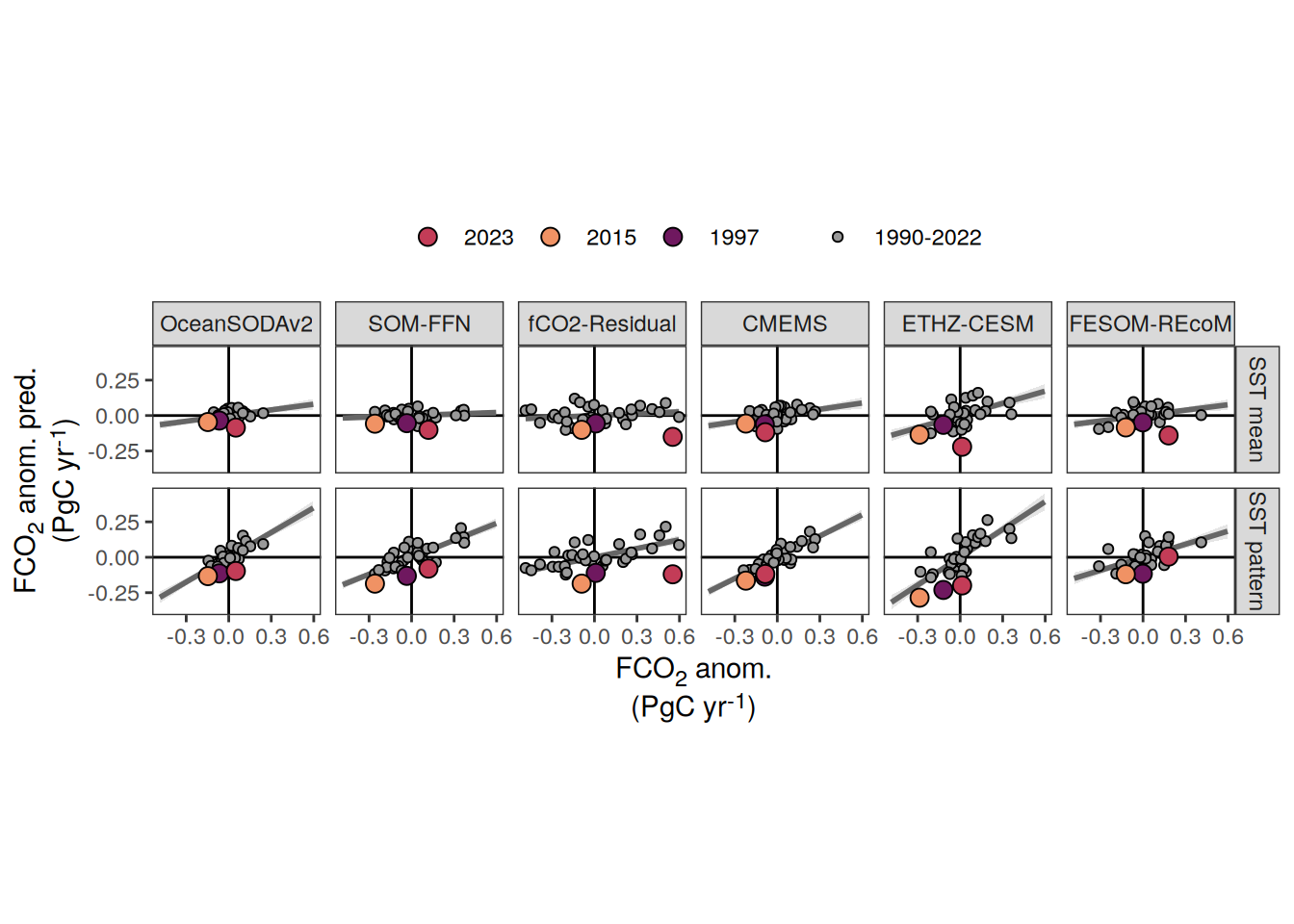
ggsave(width = 10,
height = 4,
dpi = 600,
filename = "../output/biome_flux_anomaly_prediction_correlation_all_pco2_products_global.jpg")
pco2_product_biome_annual_anomaly_temperature_predict %>%
select(product,
year,
biome,
fgco2_int,
`SST pattern` = fgco2_predict_int,
`SST mean` = fgco2_predict_int_biome) %>%
pivot_longer(contains("SST"),
values_to = "resid") %>%
filter(biome %in% c("Global non-polar")) %>%
group_by(product, biome, name) %>%
summarise(correlation = cor(fgco2_int, resid)) %>%
ungroup() %>%
ggplot(aes(name, correlation, fill = name))+
geom_hline(yintercept = 0) +
geom_col(col = "grey20") +
scale_fill_highcontrast() +
labs(y = "FCO<sub>2</sub> anom.<br>correlation") +
facet_grid( ~ product) +
theme(
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_markdown(),
legend.title = element_blank()
)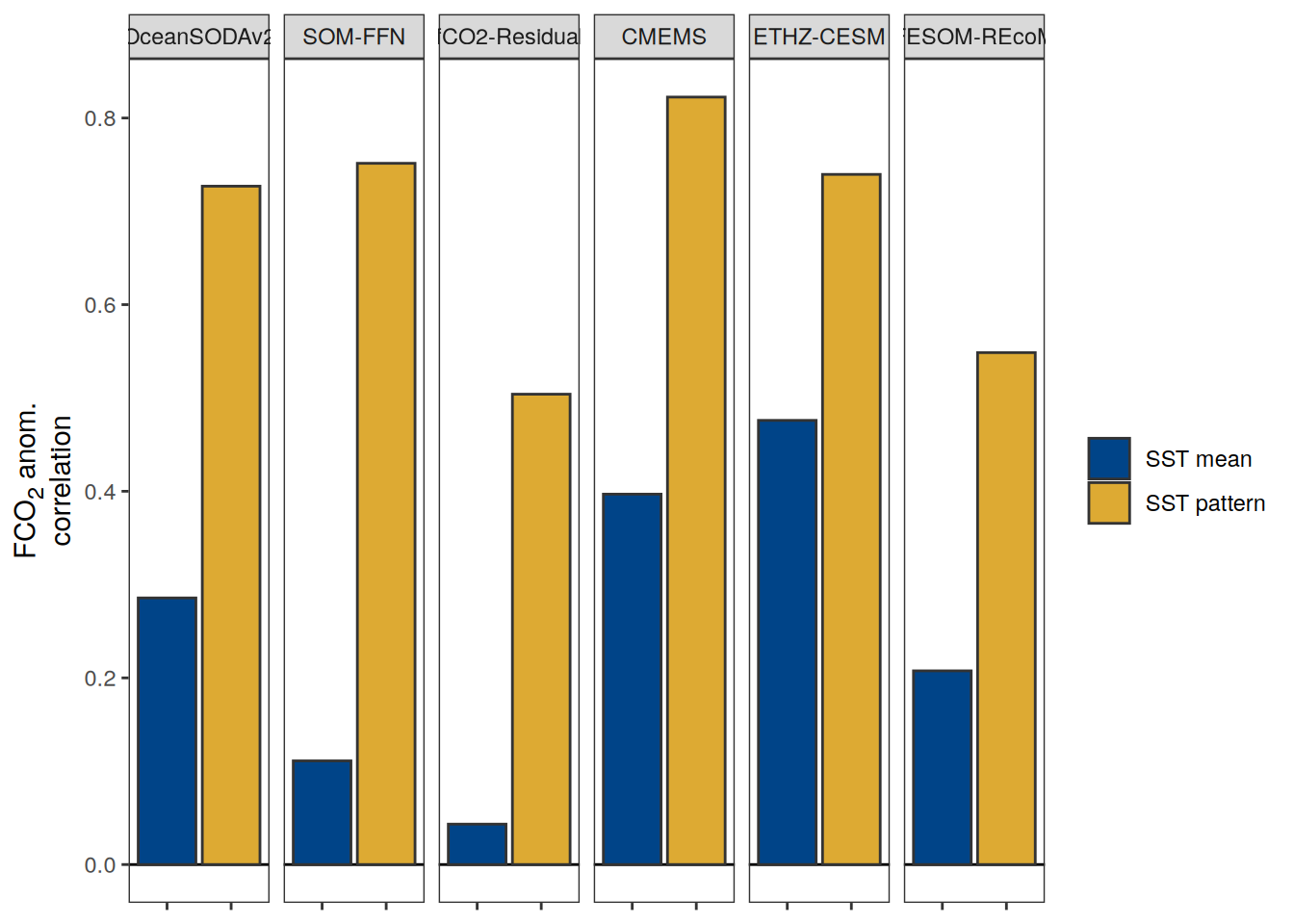
ggsave(width = 10,
height = 2,
dpi = 600,
filename = "../output/biome_flux_anomaly_prediction_correlation_coefficients_all_pco2_products_global.jpg")
pco2_product_biome_annual_anomaly_temperature_predict %>%
select(product,
year,
biome,
fgco2_int,
`SST pattern` = fgco2_predict_int) %>%
pivot_longer(contains("SST"),
values_to = "resid") %>%
filter(biome %in% c(key_biomes)) %>%
ggplot(aes(fgco2_int, resid))+
geom_vline(xintercept = 0) +
geom_hline(yintercept = 0) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_point(data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(fill = "1990-2022"),
shape = 21) +
scale_color_manual(values = "grey60", name = "X") +
scale_fill_manual(values = "grey60", name = "X") +
new_scale_fill() +
new_scale_color() +
geom_point(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(fill = as.factor(year)),
shape = 21,
size = 3
) +
scale_fill_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
scale_color_manual(values = rev(warm_cool_gradient[c(17, 13, 20)]),
guide = guide_legend(reverse = TRUE, order = 2)) +
labs(x = labels_breaks("fgco2_int")$i_legend_title,
y = labels_breaks("fgco2_predict_int")$i_legend_title) +
facet_grid(biome ~ product) +
coord_equal() +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)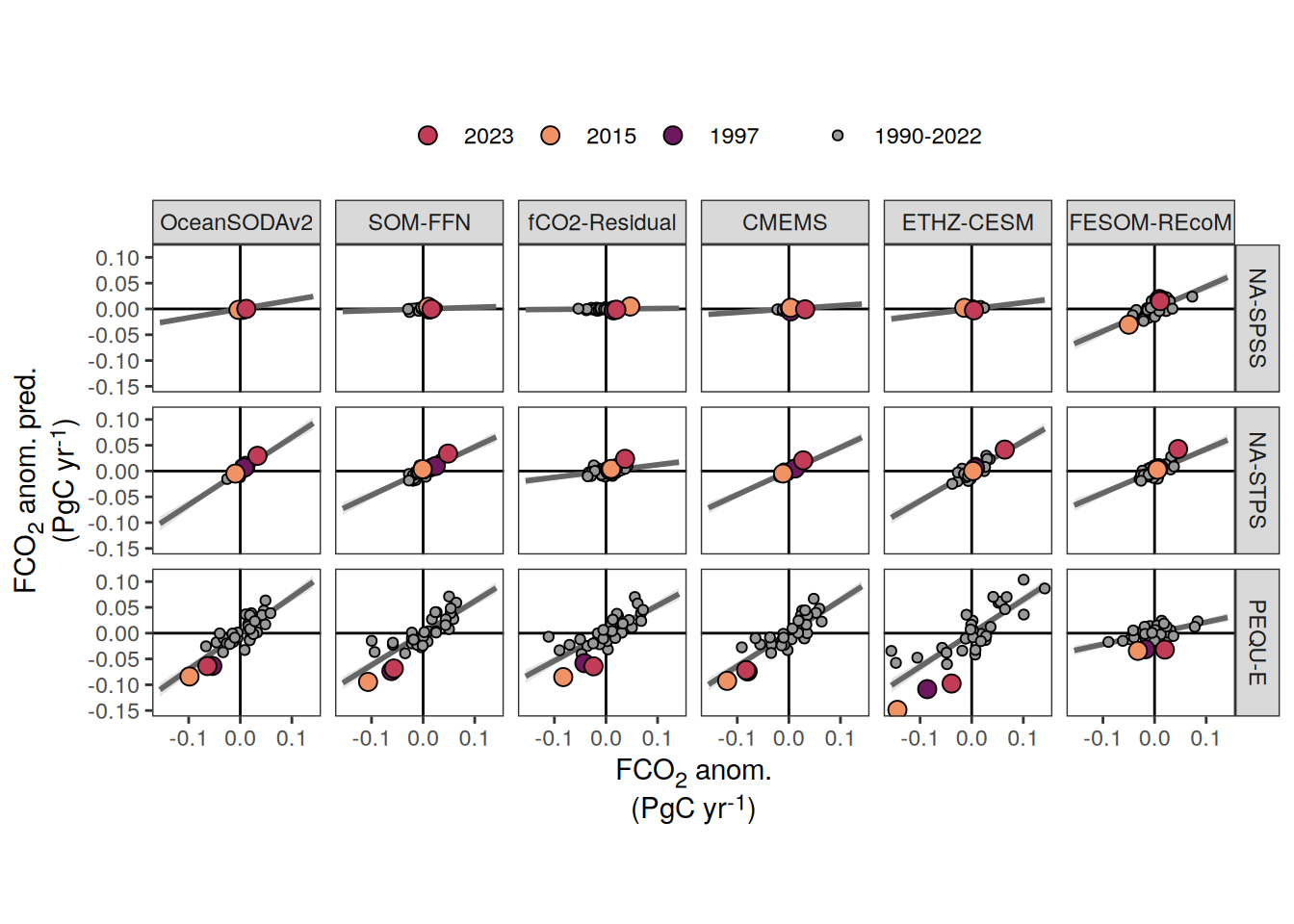
| Version | Author | Date |
|---|---|---|
| f2beca5 | jens-daniel-mueller | 2025-03-04 |
pco2_product_biome_annual_anomaly_temperature_predict %>%
select(product, year, biome, fgco2_int, fgco2_predict_int, fgco2_predict_int_biome) %>%
pivot_longer(contains("predict"),
values_to = "resid") %>%
filter(biome %in% c("Global non-polar")) %>%
ggplot(aes(fgco2_int - resid, fill = name)) +
geom_vline(xintercept = 0) +
geom_histogram(binwidth=.05, alpha=.5, position="identity") +
geom_density(aes(col = name), fill = "transparent") +
facet_wrap(~ product, scales = "free") +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)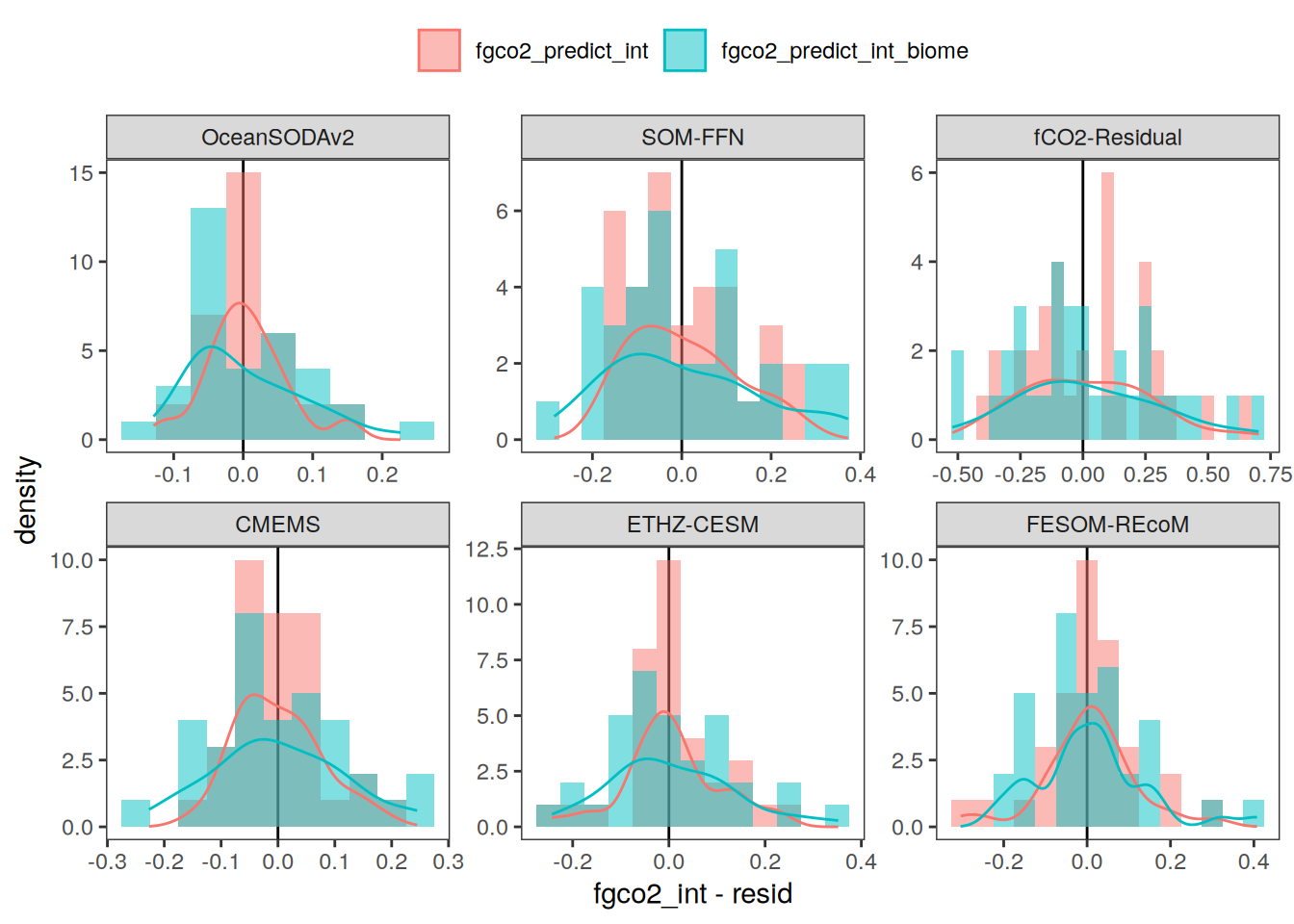
| Version | Author | Date |
|---|---|---|
| f2beca5 | jens-daniel-mueller | 2025-03-04 |
Monthly anomalies
Absolute
pco2_product_biome_monthly_detrended %>%
filter(biome == "Global non-polar") %>%
select(-c(time, fit, value)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(product, year, month, biome, fgco2_int)) %>%
filter(name == "temperature") %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2
) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(
data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3
) +
scale_color_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
scale_fill_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
labs(
y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks(unique(.x$name))$i_legend_title
) +
facet_grid(biome ~ product,
scales = "free_y") +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown()
)
)[[1]]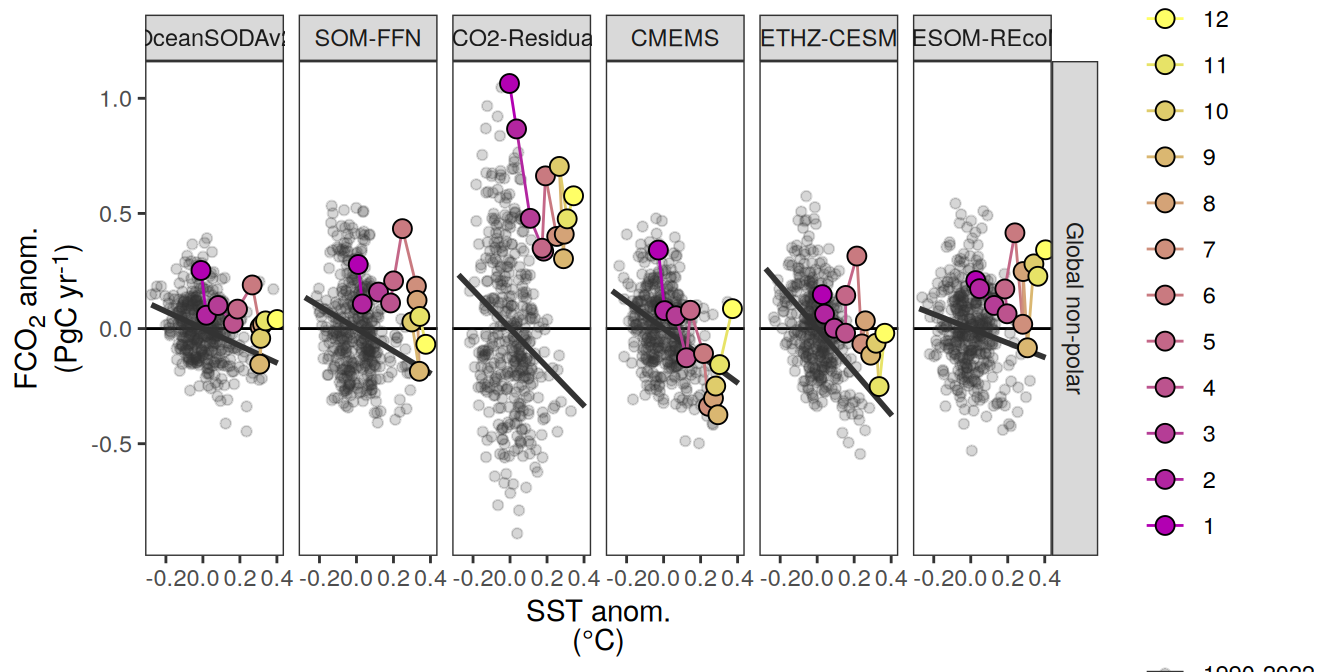
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
| 58e3680 | jens-daniel-mueller | 2024-04-11 |
pco2_product_biome_monthly_detrended %>%
filter(biome %in% key_biomes) %>%
select(-c(time, fit, value)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(product, year, month, biome, fgco2_int)) %>%
filter(name == "temperature") %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2
) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(
data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3
) +
scale_color_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
scale_fill_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
labs(
y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks(unique(.x$name))$i_legend_title
) +
facet_grid(biome ~ product,
scales = "free_y") +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown()
)
)[[1]]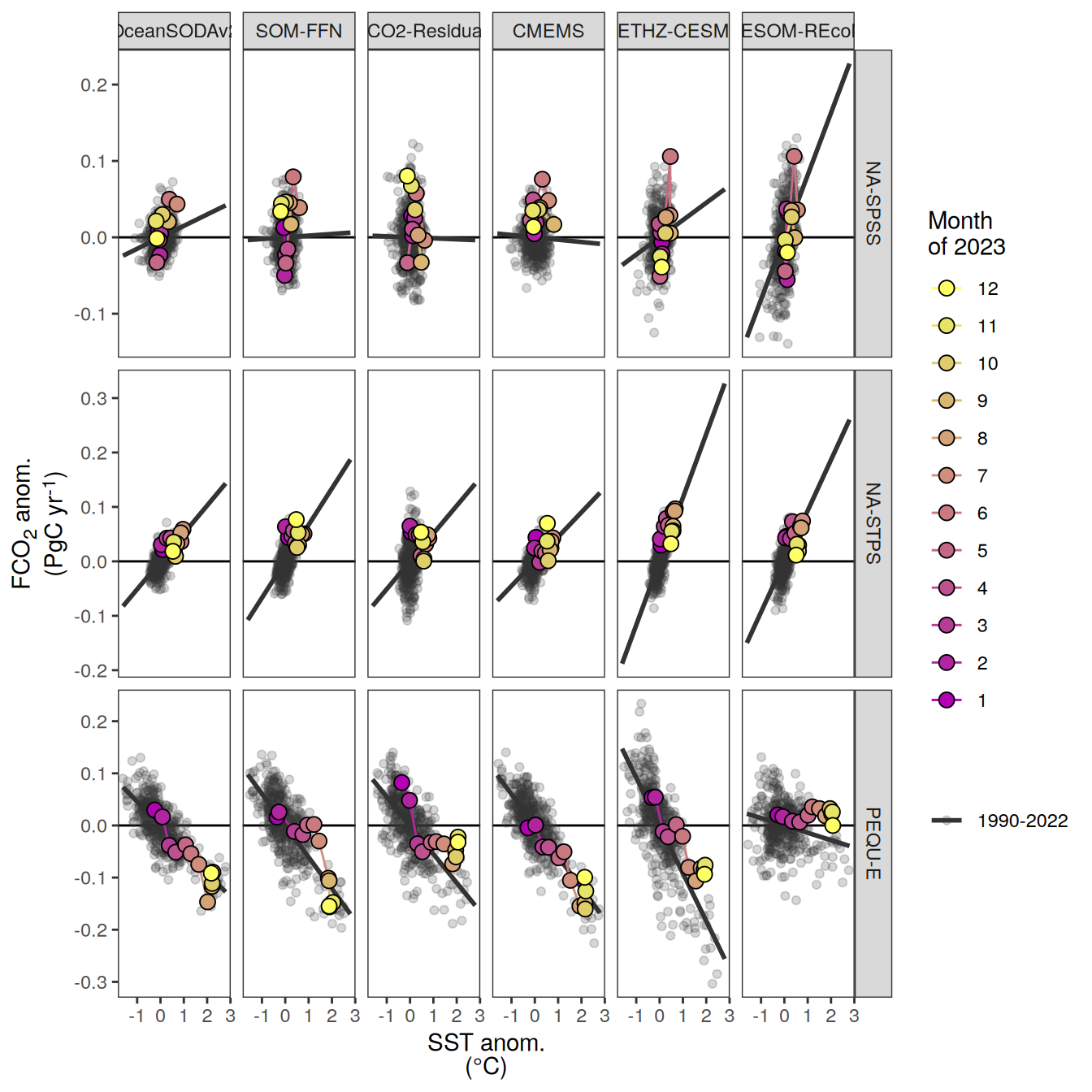
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| d82bd91 | jens-daniel-mueller | 2024-08-27 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 8c96de4 | jens-daniel-mueller | 2024-05-08 |
| 4b81eaf | jens-daniel-mueller | 2024-05-07 |
| 60abdac | jens-daniel-mueller | 2024-04-23 |
| 1ff6eb0 | jens-daniel-mueller | 2024-04-22 |
| 9ecd92e | jens-daniel-mueller | 2024-04-22 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| a5911f0 | jens-daniel-mueller | 2024-04-17 |
| 6709afa | jens-daniel-mueller | 2024-04-12 |
fCO2 decomposition
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(biome %in% c("Global non-polar",key_biomes)) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ p_season(df = .x,
title = paste("Anomalies from predicted monthly mean |", .x$biome))
)[[1]]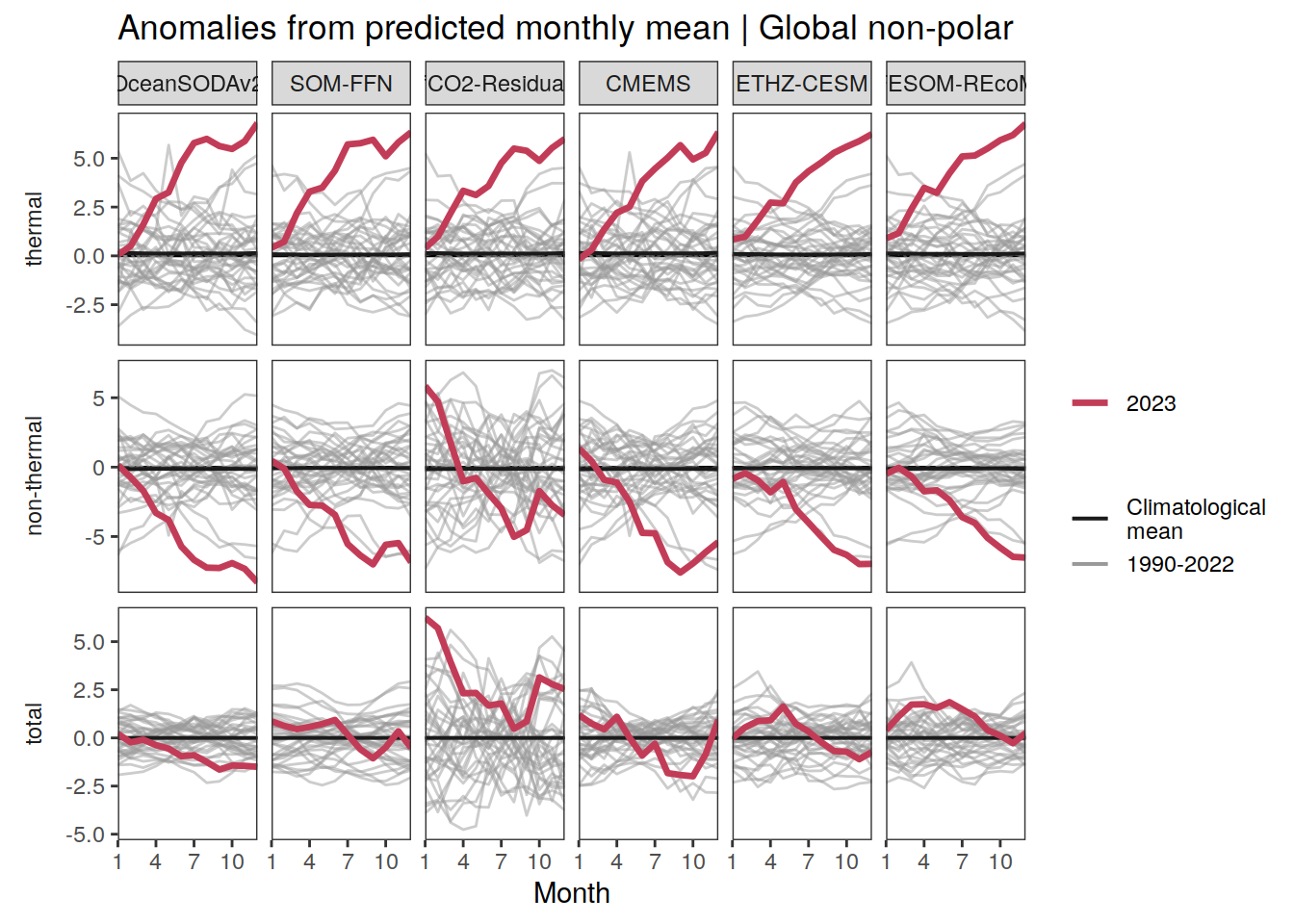
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
[[2]]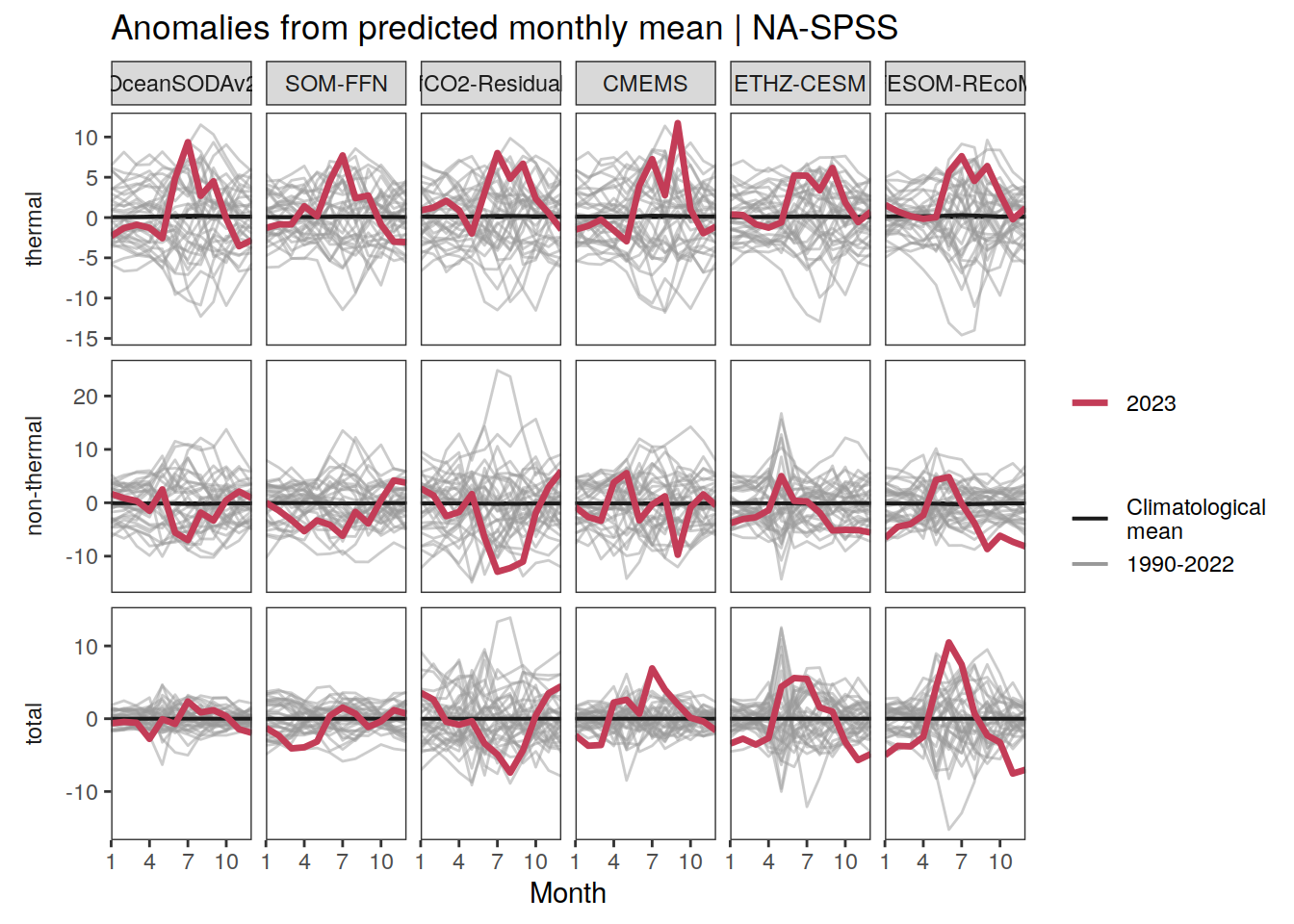
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
[[3]]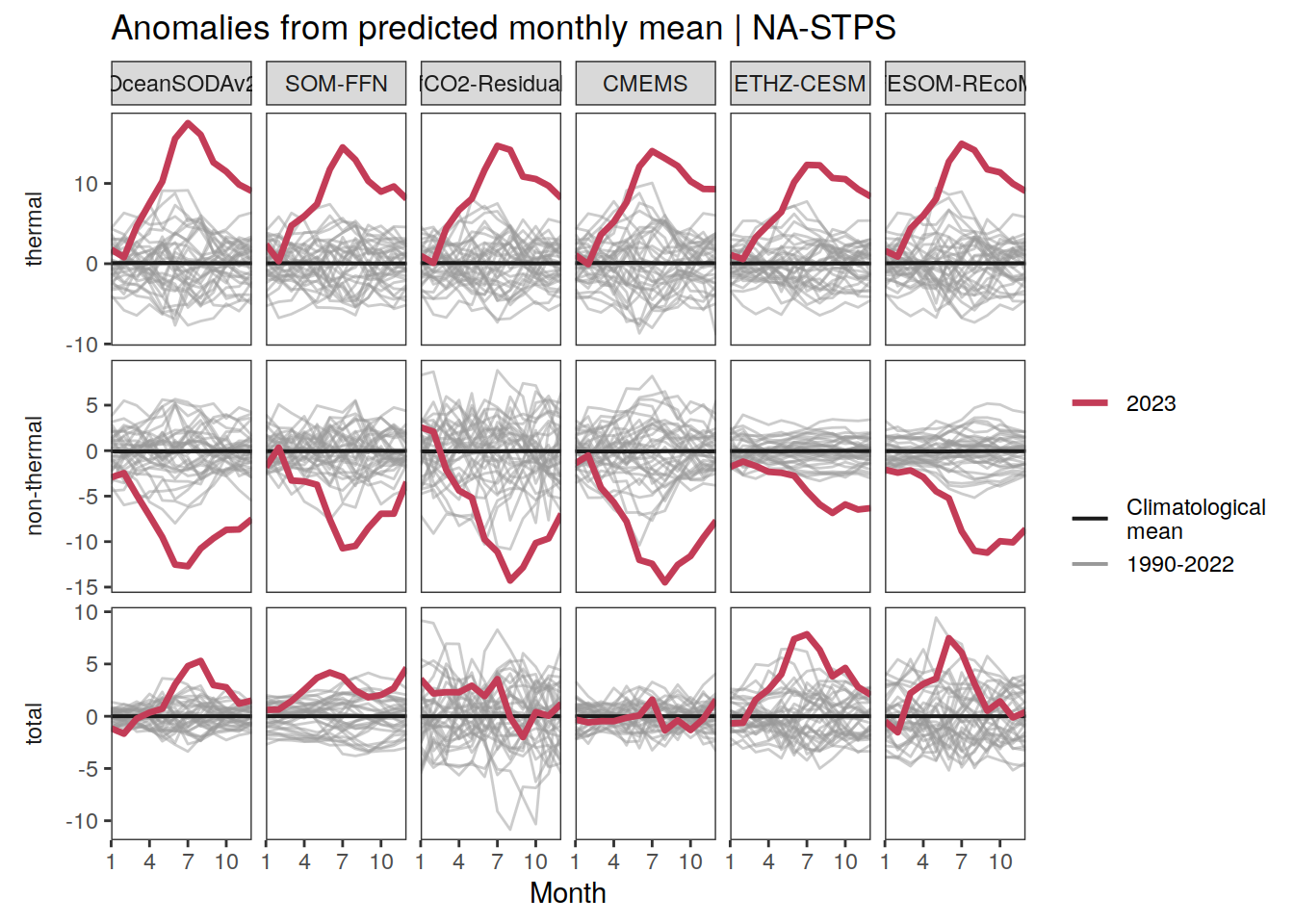
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
[[4]]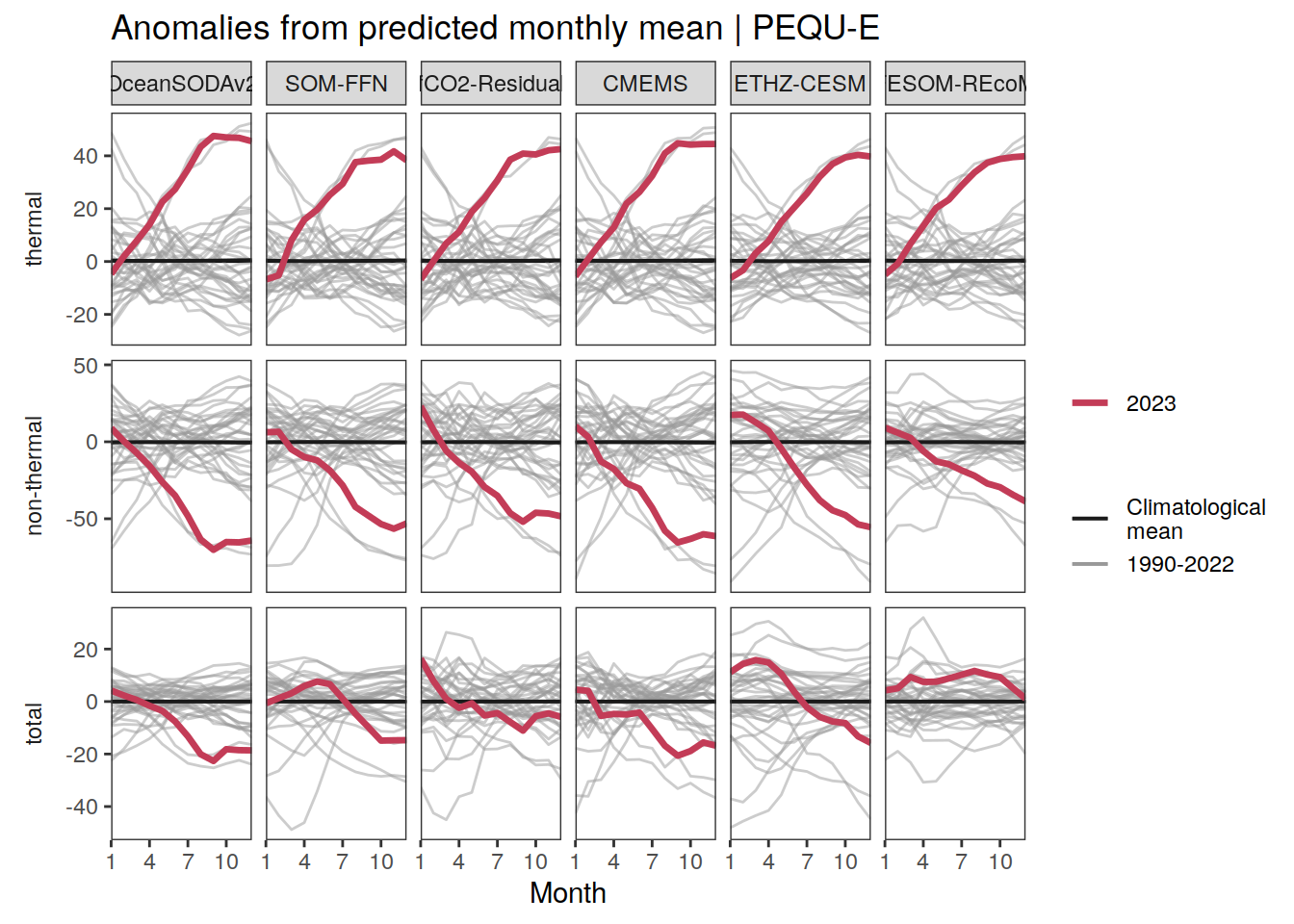
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 5af03d1 | jens-daniel-mueller | 2024-05-17 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| 1e4c153 | jens-daniel-mueller | 2024-05-14 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
pco2_product_biome_annual_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(product %in% pco2_product_list) %>%
group_by(year, name, biome, product) %>%
summarise(resid = mean(resid)) %>%
ungroup() %>%
group_by(year, name, biome) %>%
summarise(resid_sd = sd(resid), resid = mean(resid)) %>%
ungroup()
pco2_product_biome_annual_fCO2_decomposition %>%
ggplot(aes(year, resid, colour = name)) +
geom_hline(yintercept = 0) +
geom_path() +
facet_wrap( ~ biome)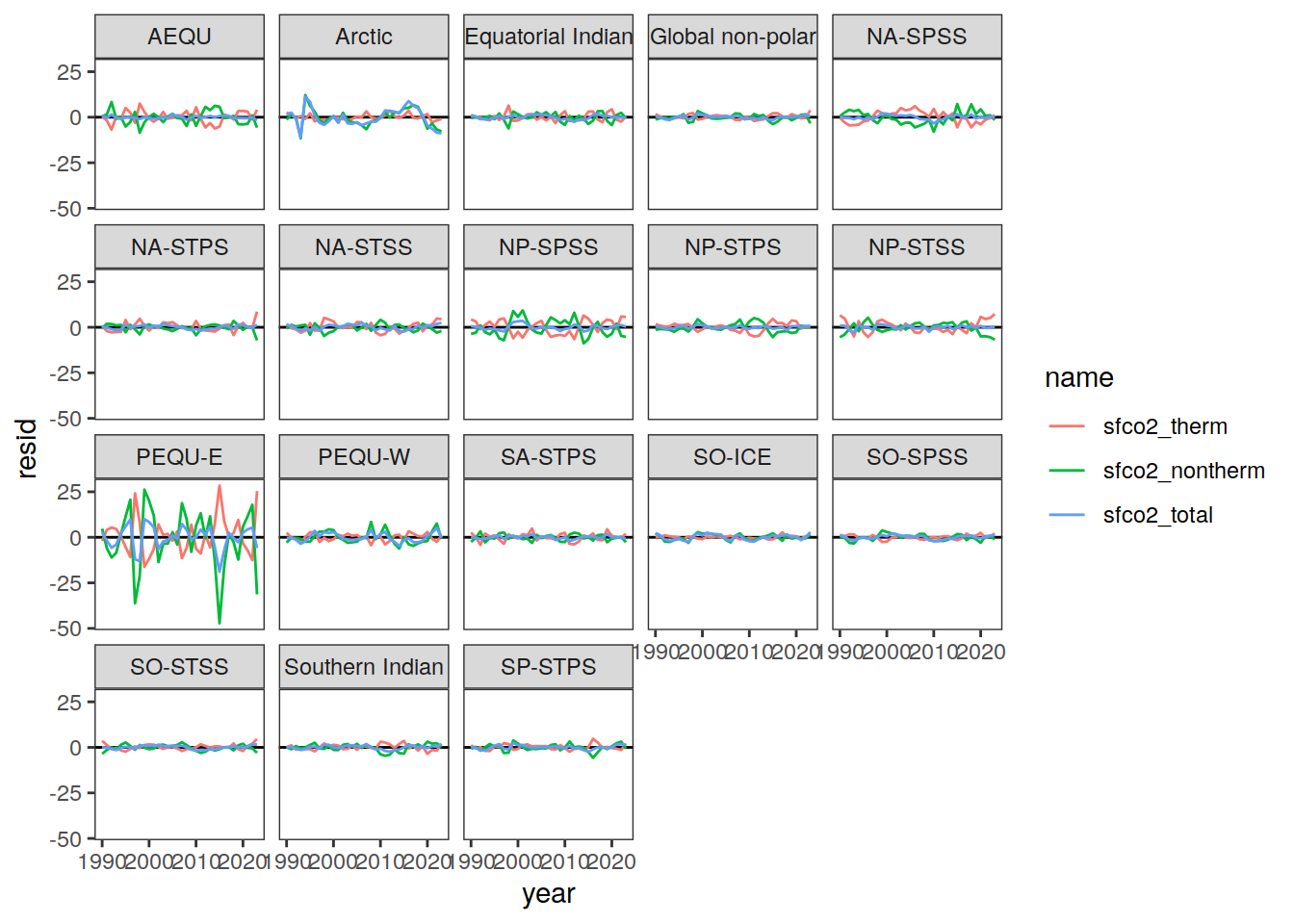
pco2_product_biome_annual_fCO2_decomposition %>%
pivot_wider(values_from = contains("resid")) %>%
ggplot(aes(resid_sfco2_therm, resid_sfco2_nontherm, col = "observed")) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 0) +
geom_abline(slope = -1, intercept = 0) +
geom_smooth(method = "lm", se = FALSE) +
geom_point(shape = 21) +
scale_color_muted() +
facet_wrap( ~ biome, scales = "free")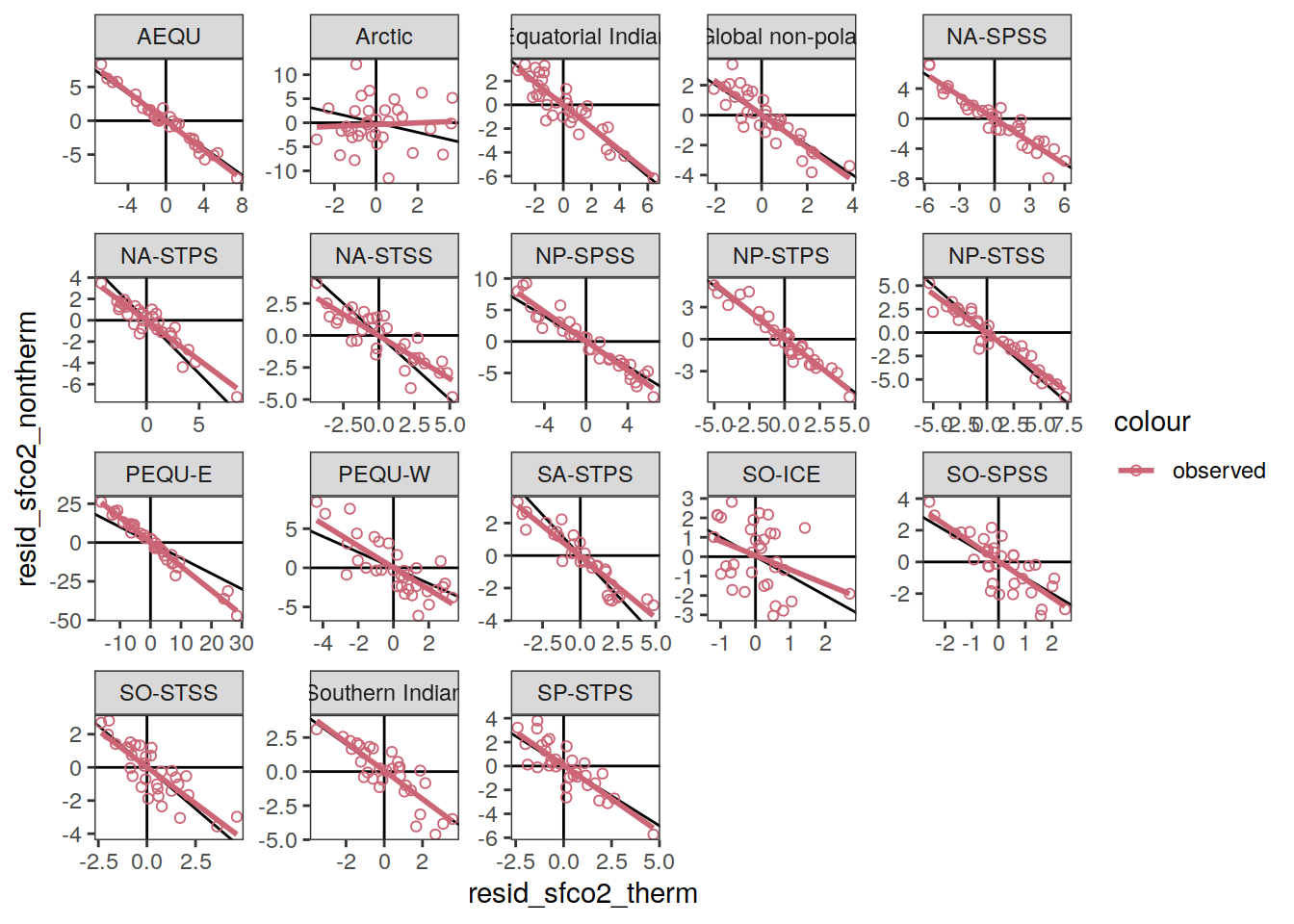
pco2_product_biome_annual_fCO2_decomposition %>%
filter(year == 2023) %>%
ggplot(aes(name, resid, fill = name)) +
geom_hline(yintercept = 0) +
geom_col(col = "grey20") +
scale_fill_manual(values = c(warm_color, cold_color, "grey80")) +
labs(y = labels_breaks("sfco2")$i_legend_title) +
facet_wrap(~ biome, scales = "free_y") +
theme(
legend.title = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
legend.position = c(0.9, 0.1)
)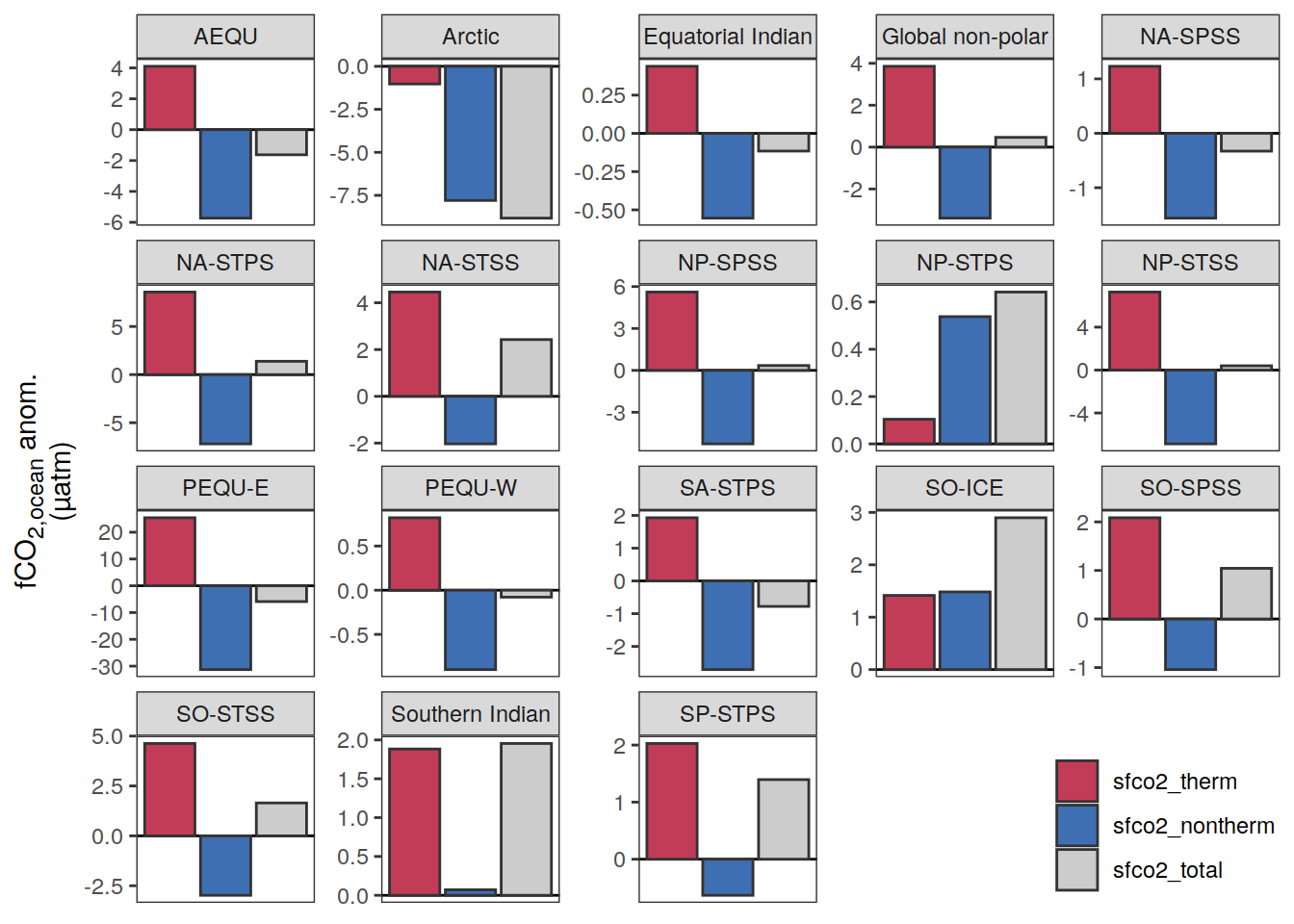
pco2_product_biome_annual_fCO2_decomposition %>%
filter(year == 2023, biome %in% c("PEQU-E", "NA-STPS")) %>%
mutate(name = case_when(
name == "sfco2_therm" ~ "thermal",
name == "sfco2_nontherm" ~ "non-thermal",
name == "sfco2_total" ~ "total"
),
name = fct_inorder(name)) %>%
ggplot(aes(name, resid, fill = name)) +
geom_hline(yintercept = 0) +
geom_col(col = "grey20") +
geom_text(
data = . %>% filter(biome == "NA-STPS"),aes(
label = name,
col = name,
hjust = if_else(sign(resid) > 0, 0, 1),
y = resid + if_else(sign(resid) > 0, 1, -1)
),
angle = 90,
fontface = "bold") +
scale_color_manual(values = c(warm_color, cold_color, "grey20")) +
scale_fill_manual(values = c(warm_color, cold_color, "grey20")) +
labs(y = labels_breaks("sfco2")$i_legend_title) +
scale_y_continuous(breaks = seq(-20, 20, 20)) +
facet_grid(. ~ fct_rev(biome)) +
theme_classic() +
theme(
legend.title = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
strip.background = element_blank(),
strip.text = element_text(face = "bold", size = 16),
axis.line.x = element_blank(),
legend.position = "none"
)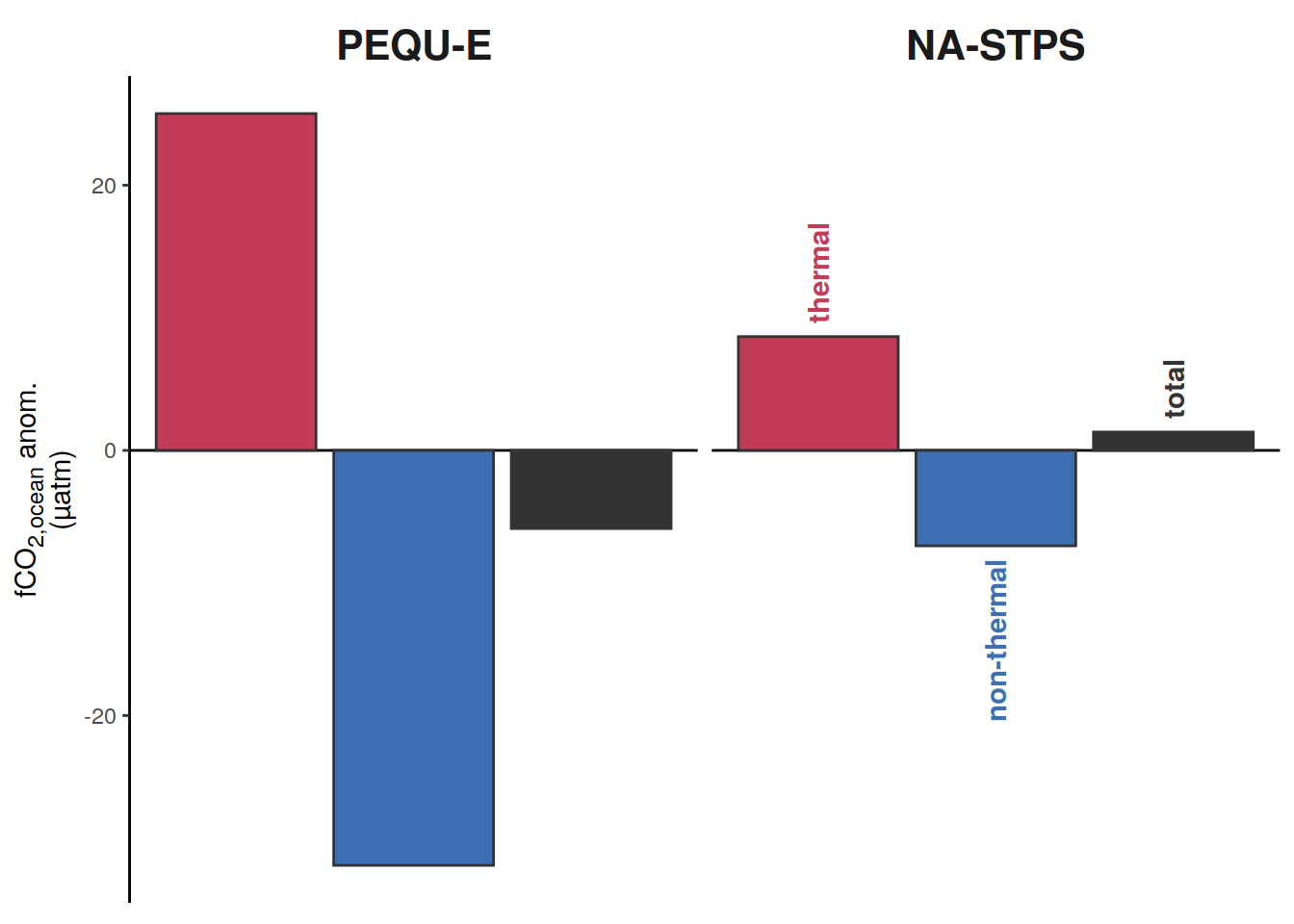
# ggsave(width = 6,
# height = 3,
# dpi = 600,
# filename = "../output/biome_annual_fco2_decomposition.jpg")
pco2_product_biome_annual_fCO2_decomposition %>%
filter(year == 2023) %>%
mutate(name = case_when(
name == "sfco2_therm" ~ "thermal",
name == "sfco2_nontherm" ~ "non-thermal",
name == "sfco2_total" ~ "total"
),
name = fct_inorder(name)) %>%
ggplot(aes(name, resid, fill = name)) +
geom_hline(yintercept = 0) +
geom_col(col = "grey20") +
geom_linerange(aes(
name,
ymin = resid - resid_sd,
ymax = resid + resid_sd
), col = "grey20") +
scale_color_manual(values = c(warm_color, cold_color, "grey20")) +
scale_fill_manual(values = c(warm_color, cold_color, "grey20")) +
labs(y = labels_breaks("sfco2")$i_legend_title) +
facet_wrap(. ~ biome, scales = "free_y", ncol = 4) +
theme(
legend.title = element_blank(),
legend.position = c(0.9,0.1),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_markdown()
)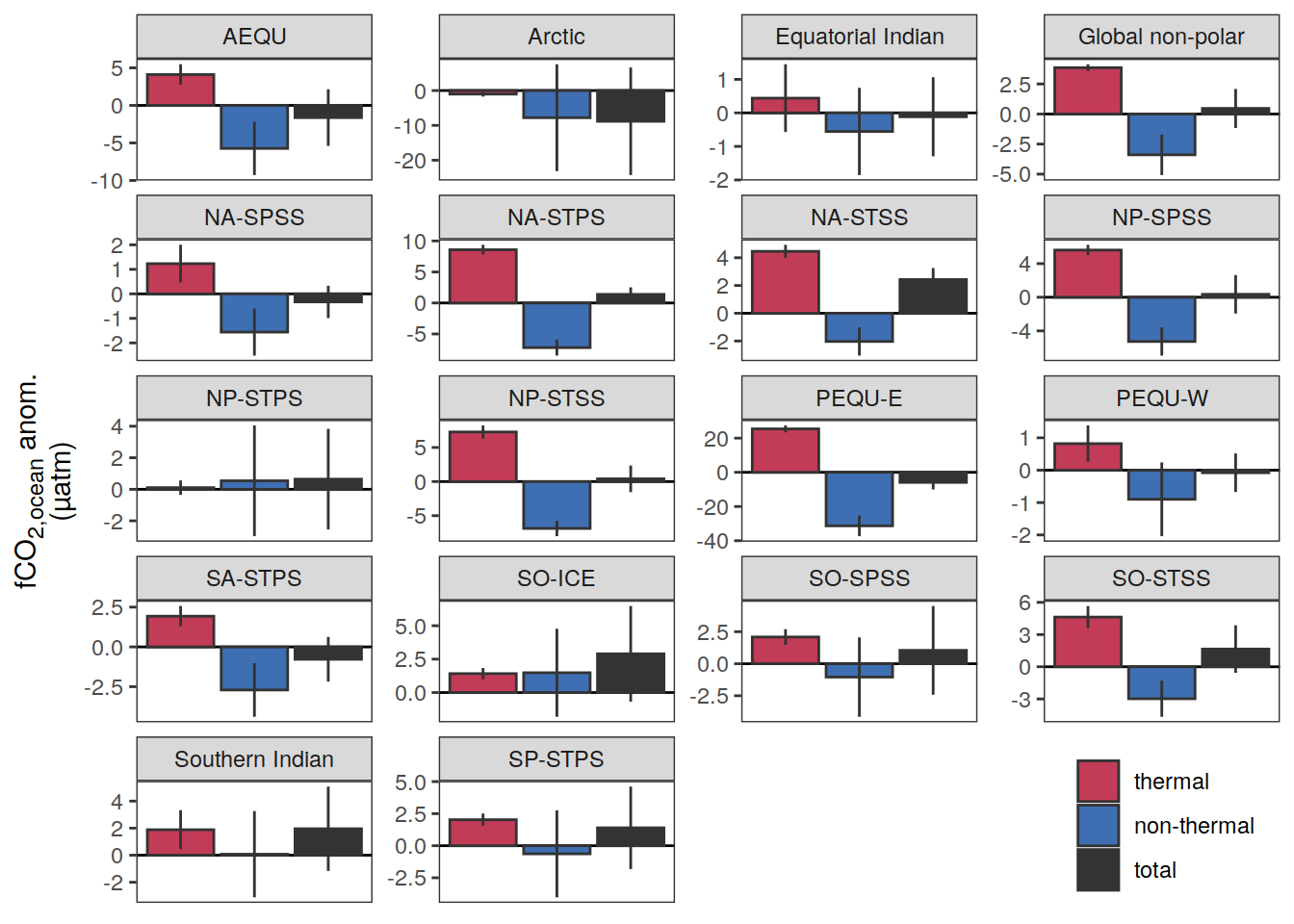
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 713d232 | jens-daniel-mueller | 2024-09-12 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
ggsave(width = 7,
height = 7,
dpi = 600,
filename = "../output/biome_annual_fco2_decomposition_all_biomes.jpg")Flux attribution
Seasonal
pco2_product_biome_annual_flux_attribution_ensemble <-
pco2_product_biome_annual_flux_attribution %>%
filter(product %in% pco2_product_list) %>%
group_by(biome, name) %>%
summarise(
resid_sd = sd(resid),
resid = mean(resid)) %>%
ungroup()
pco2_product_biome_annual_flux_attribution_ensemble %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_col(aes("", resid), fill = "grey90", col = "grey20") +
geom_point(
data = pco2_product_biome_annual_flux_attribution %>%
filter(biome %in% c("Global non-polar", key_biomes)),
aes("", resid, fill = product),
shape = 21
) +
scale_fill_manual(values = color_products) +
scale_y_continuous(breaks = seq(-10, 10, 0.1)) +
labs(y = labels_breaks(unique("fgco2"))$i_legend_title) +
facet_grid(
biome ~ name,
labeller = labeller(name = x_axis_labels),
scales = "free_y",
space = "free_y",
switch = "x"
) +
theme(
legend.title = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
legend.position = "top"
)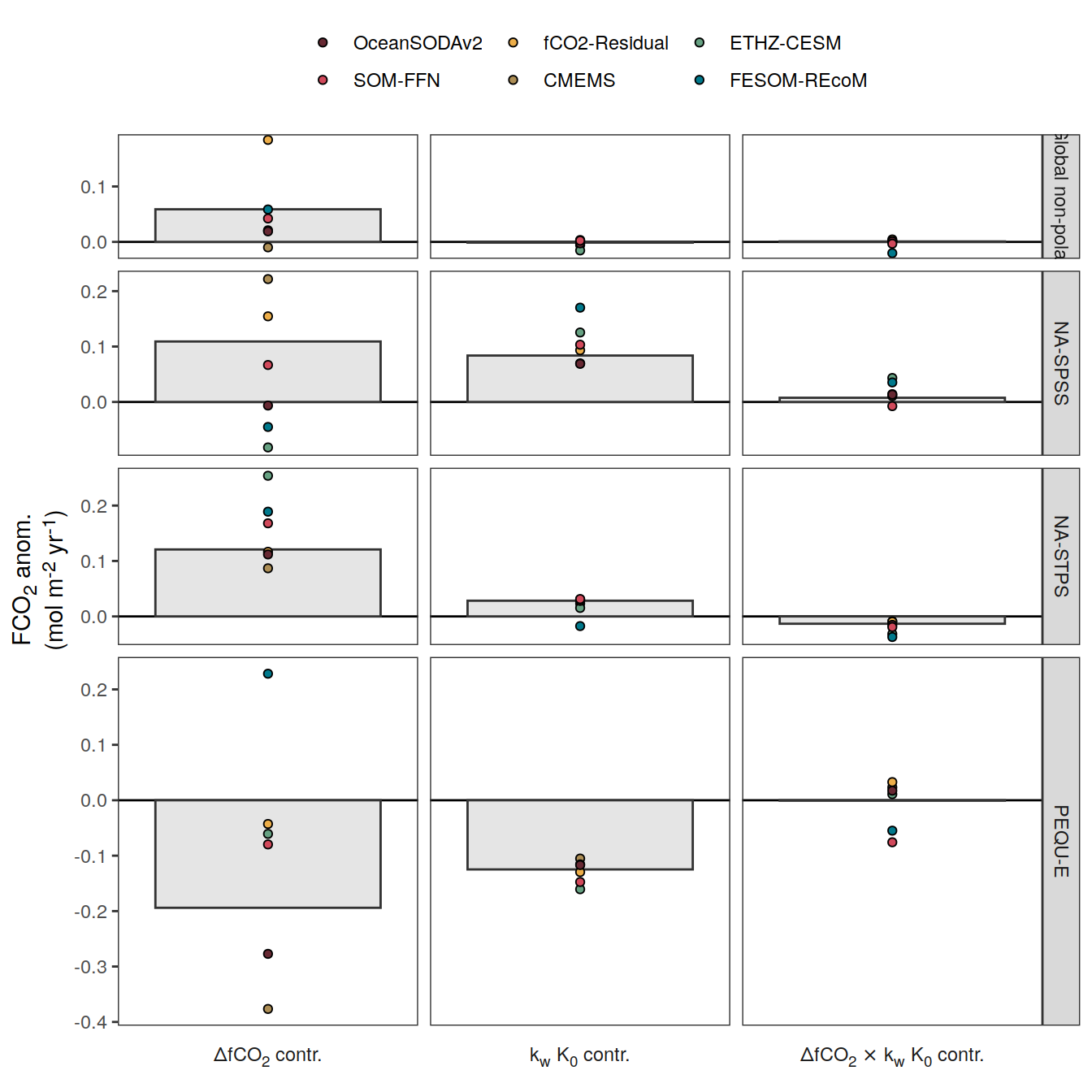
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 563345f | jens-daniel-mueller | 2024-05-21 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
| 00ad9d5 | jens-daniel-mueller | 2024-05-15 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
pco2_product_biome_annual_flux_attribution_ensemble %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_col(aes(name, resid, fill = name), col = "grey20") +
geom_linerange(aes(
name,
ymin = resid - resid_sd,
ymax = resid + resid_sd
), col = "grey20") +
scale_fill_bright(labels = x_axis_labels) +
labs(y = labels_breaks(unique("fgco2"))$i_legend_title) +
facet_wrap( ~ biome, scales = "free_y", ncol = 4) +
theme(
legend.title = element_blank(),
legend.text = element_markdown(),
legend.position = c(0.8,0.1),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_markdown()
)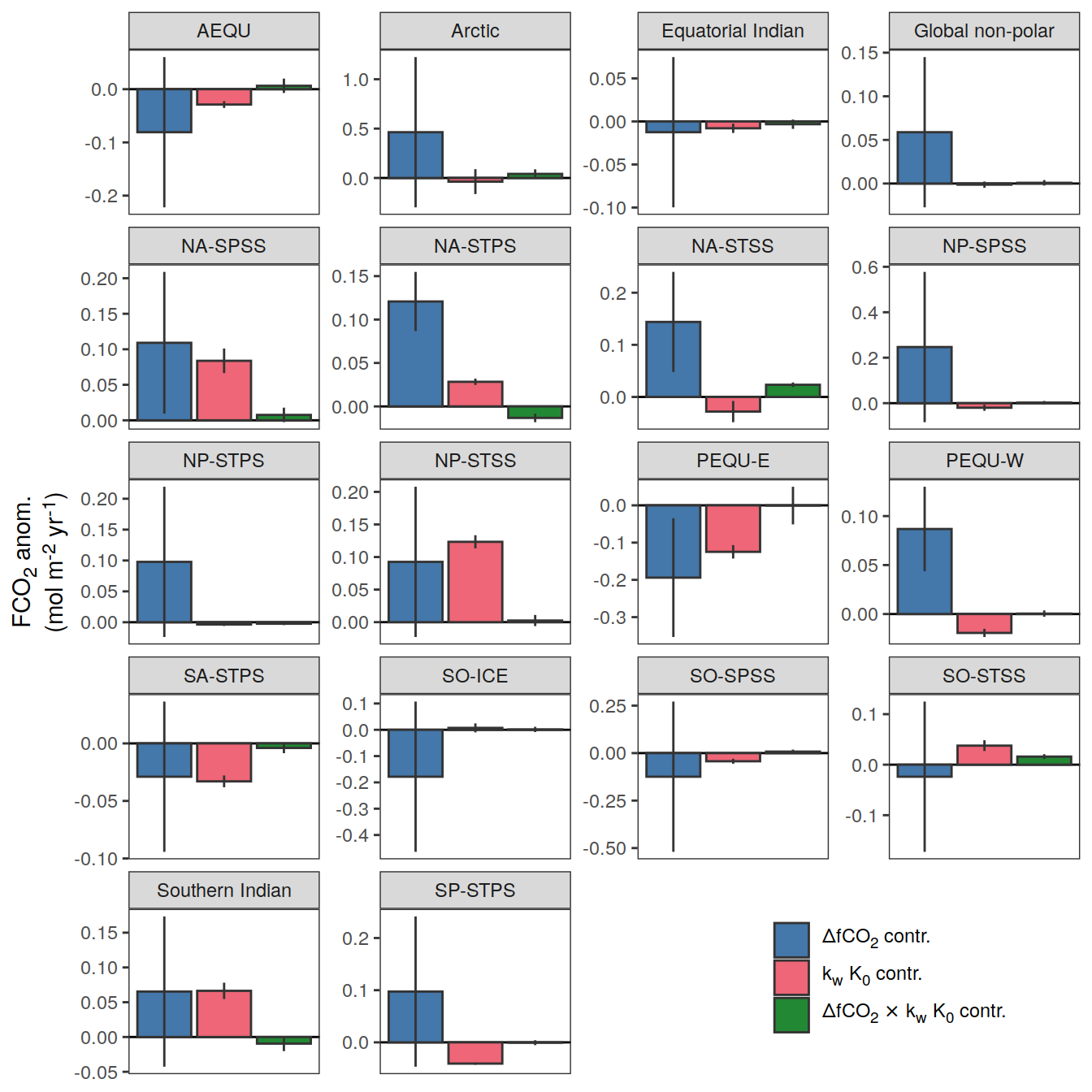
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 713d232 | jens-daniel-mueller | 2024-09-12 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 563345f | jens-daniel-mueller | 2024-05-21 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
| 00ad9d5 | jens-daniel-mueller | 2024-05-15 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
ggsave(width = 7,
height = 7,
dpi = 600,
filename = "../output/biome_annual_flux_attribution_all_biomes.jpg")
ggplot() +
geom_hline(yintercept = 0) +
geom_col(
data = pco2_product_biome_annual_flux_attribution %>%
filter(biome %in% c("Global non-polar", key_biomes)),
aes("", resid, fill = product),
position = position_dodge(width = 1),
alpha = 0.5, col = "grey30"
) +
geom_point(
data = pco2_product_biome_monthly_flux_attribution %>%
filter(year == 2023,
biome %in% c("Global non-polar", key_biomes)),
aes("", resid, fill = product),
position = position_dodge(width = 1),
shape = 21, alpha = 0.5, col = "grey30"
) +
scale_fill_manual(values = color_products) +
# scale_color_manual(values = color_products) +
scale_y_continuous(breaks = seq(-10,10,0.2)) +
labs(y = labels_breaks(unique("fgco2"))$i_legend_title) +
facet_grid(biome ~ name,
labeller = labeller(name = x_axis_labels),
scales = "free_y",
space = "free_y",
switch = "x") +
theme(
legend.title = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
legend.position = "top"
)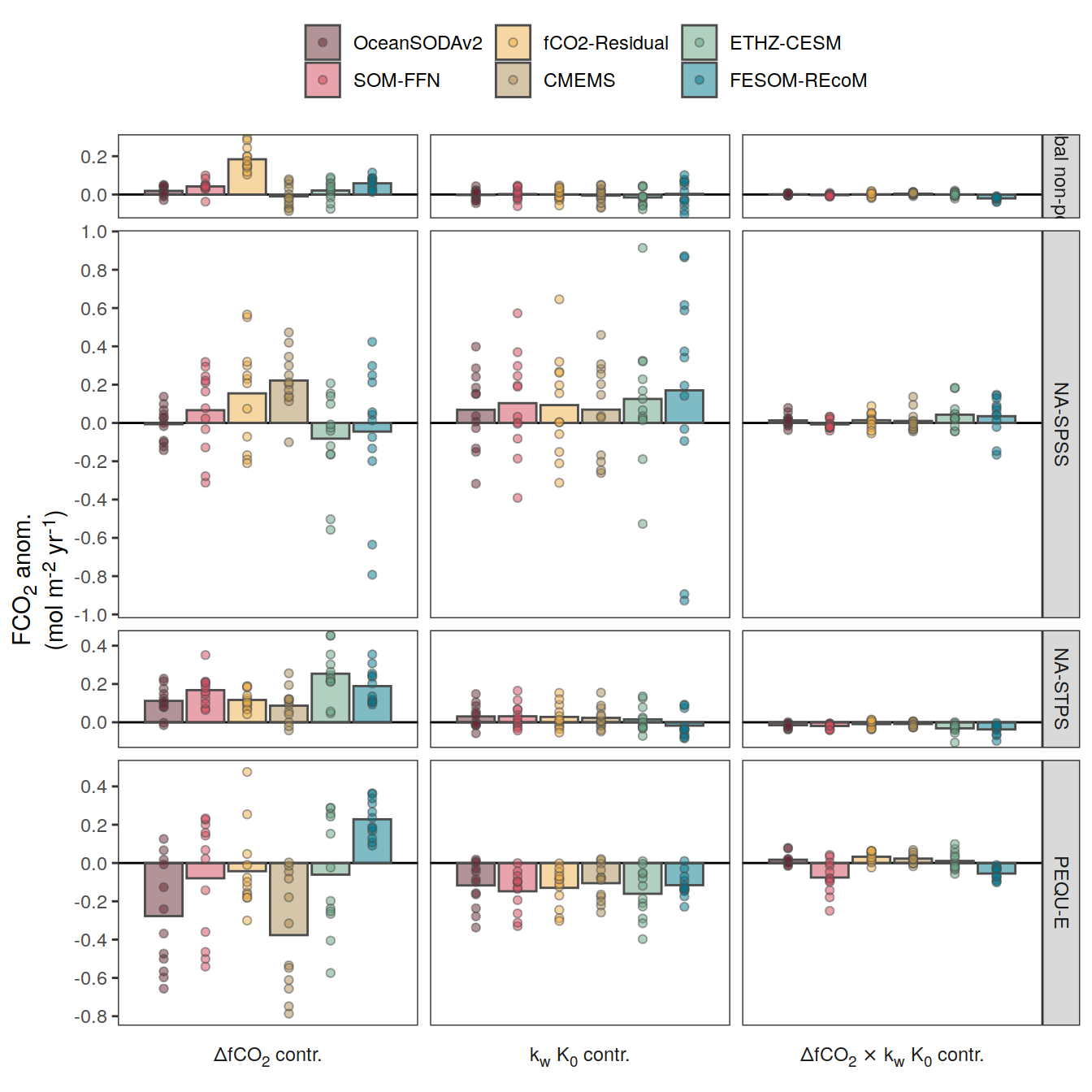
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 571e2f8 | jens-daniel-mueller | 2024-05-22 |
| 563345f | jens-daniel-mueller | 2024-05-21 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
| 00ad9d5 | jens-daniel-mueller | 2024-05-15 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
pco2_product_biome_monthly_flux_attribution %>%
filter(year == 2023,
biome %in% c("Global non-polar", key_biomes)) %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_path(
aes(month, resid, col = product)
) +
geom_point(
aes(month, resid, fill = product),
shape = 21,
alpha = 0.5,
col = "grey30"
) +
scale_fill_manual(values = color_products) +
scale_color_manual(values = color_products) +
scale_y_continuous(breaks = seq(-10,10,0.2)) +
scale_x_continuous(position = "top", breaks = seq(1,12,3)) +
labs(y = labels_breaks(unique("fgco2"))$i_legend_title) +
facet_grid(biome ~ name,
labeller = labeller(name = x_axis_labels),
scales = "free_y",
space = "free_y",
switch = "x") +
theme(
legend.title = element_blank(),
axis.title.y = element_markdown(),
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
legend.position = "top"
)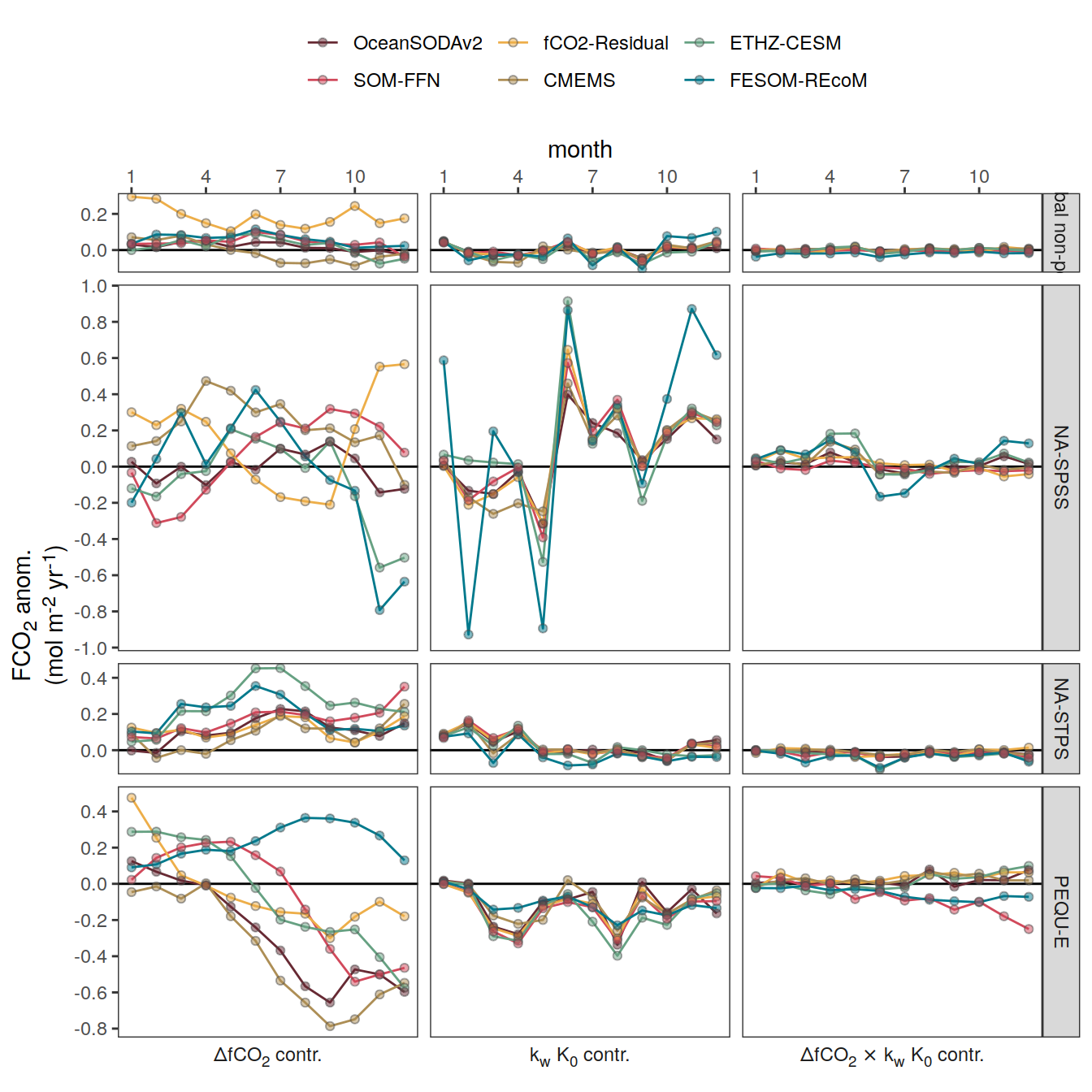
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
| 00ad9d5 | jens-daniel-mueller | 2024-05-15 |
| 47f8868 | jens-daniel-mueller | 2024-05-15 |
pco2_product_biome_monthly_flux_attribution %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ p_season(
df = .x,
title = paste("Anomalies from predicted monthly mean |", .x$biome)
) +
facet_grid(
name ~ product,
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)
)[[1]]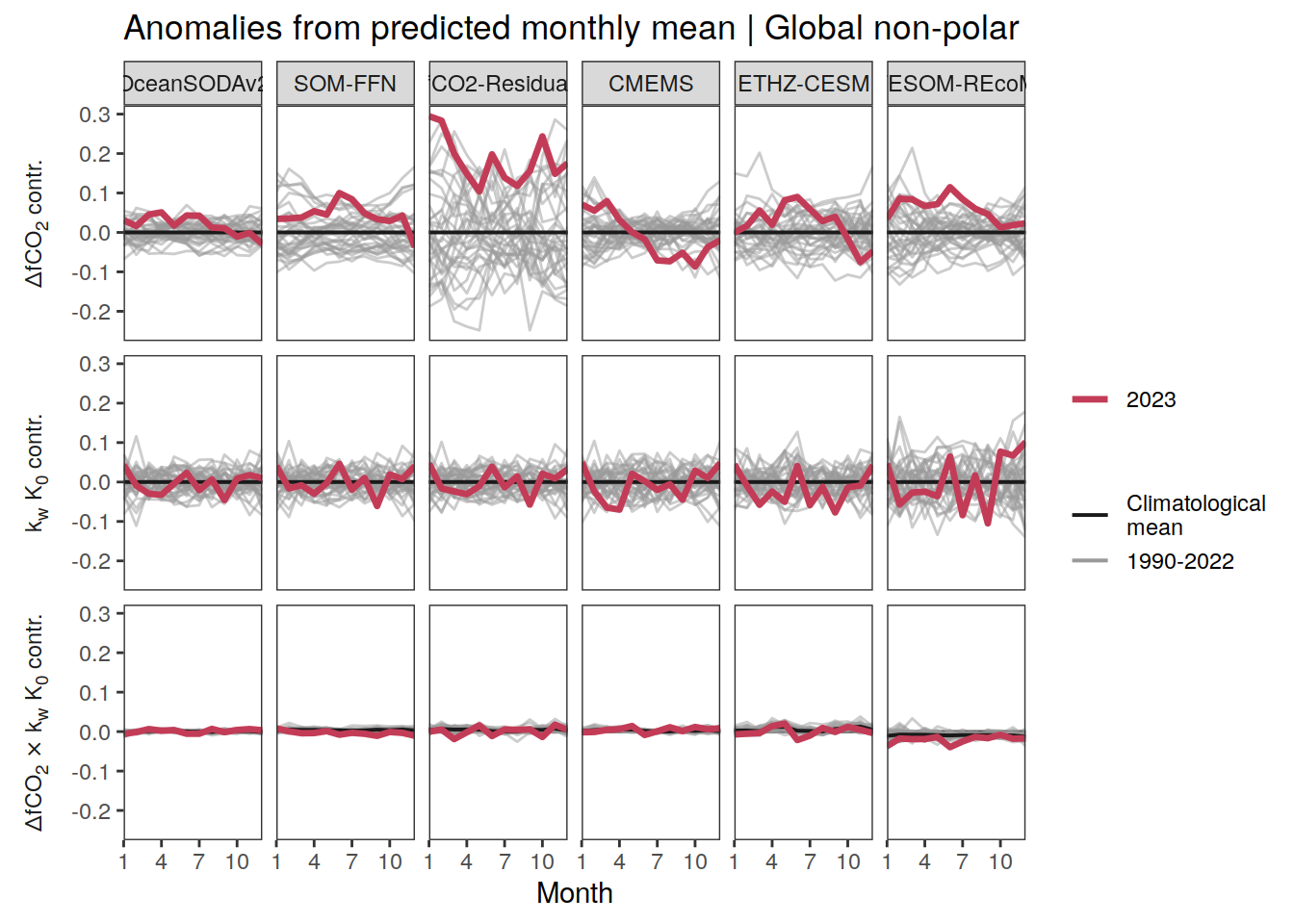
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
[[2]]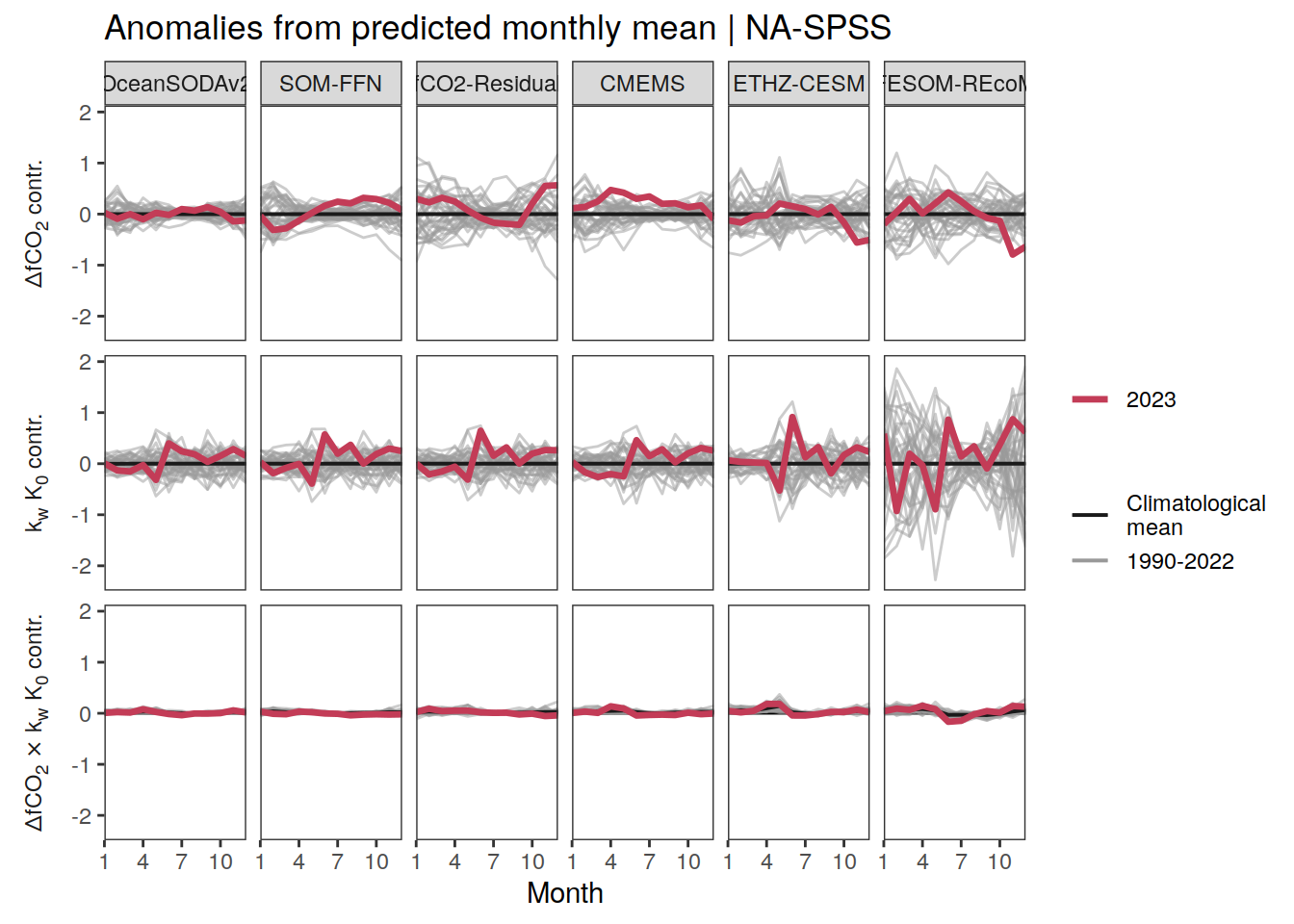
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
[[3]]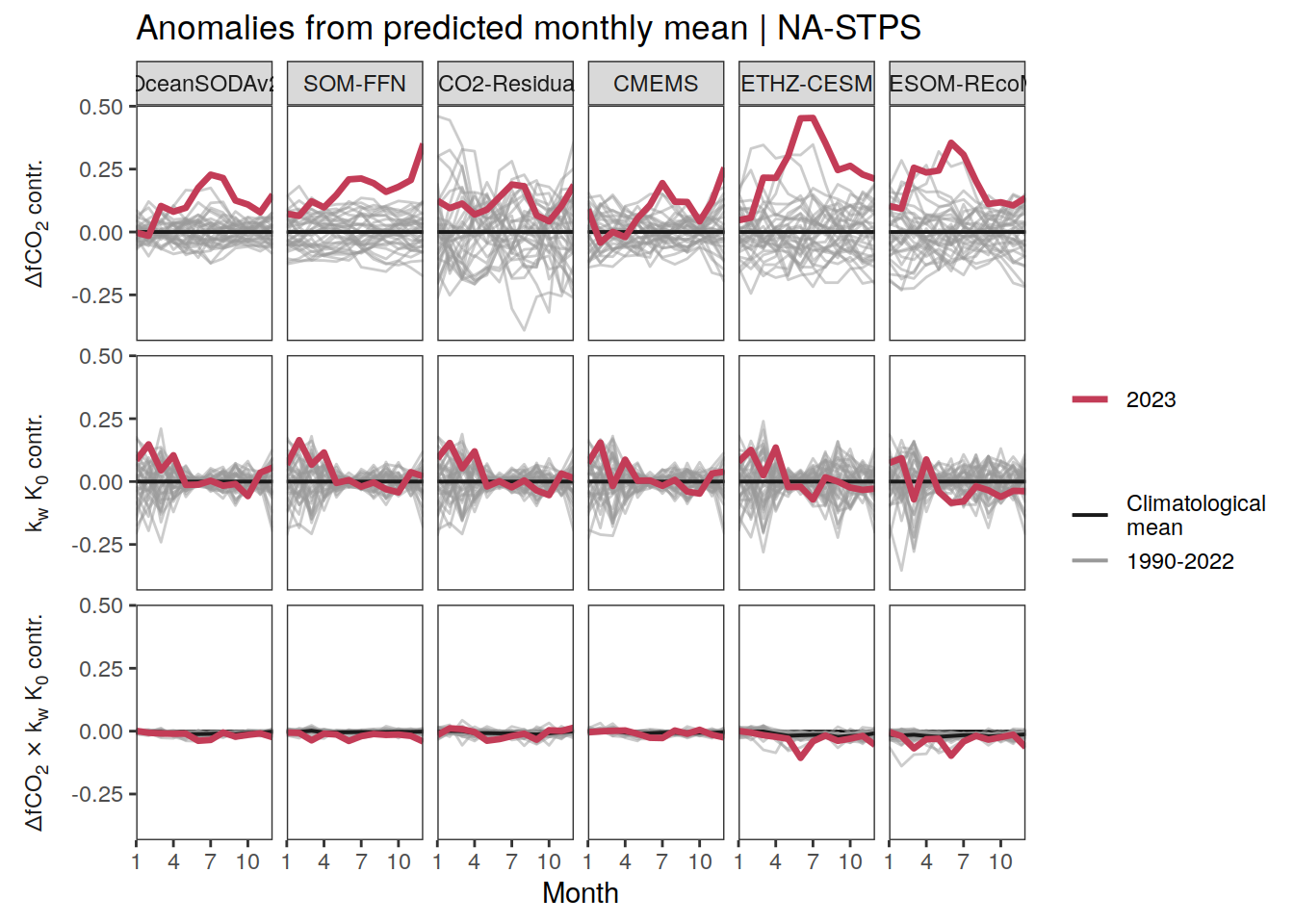
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
[[4]]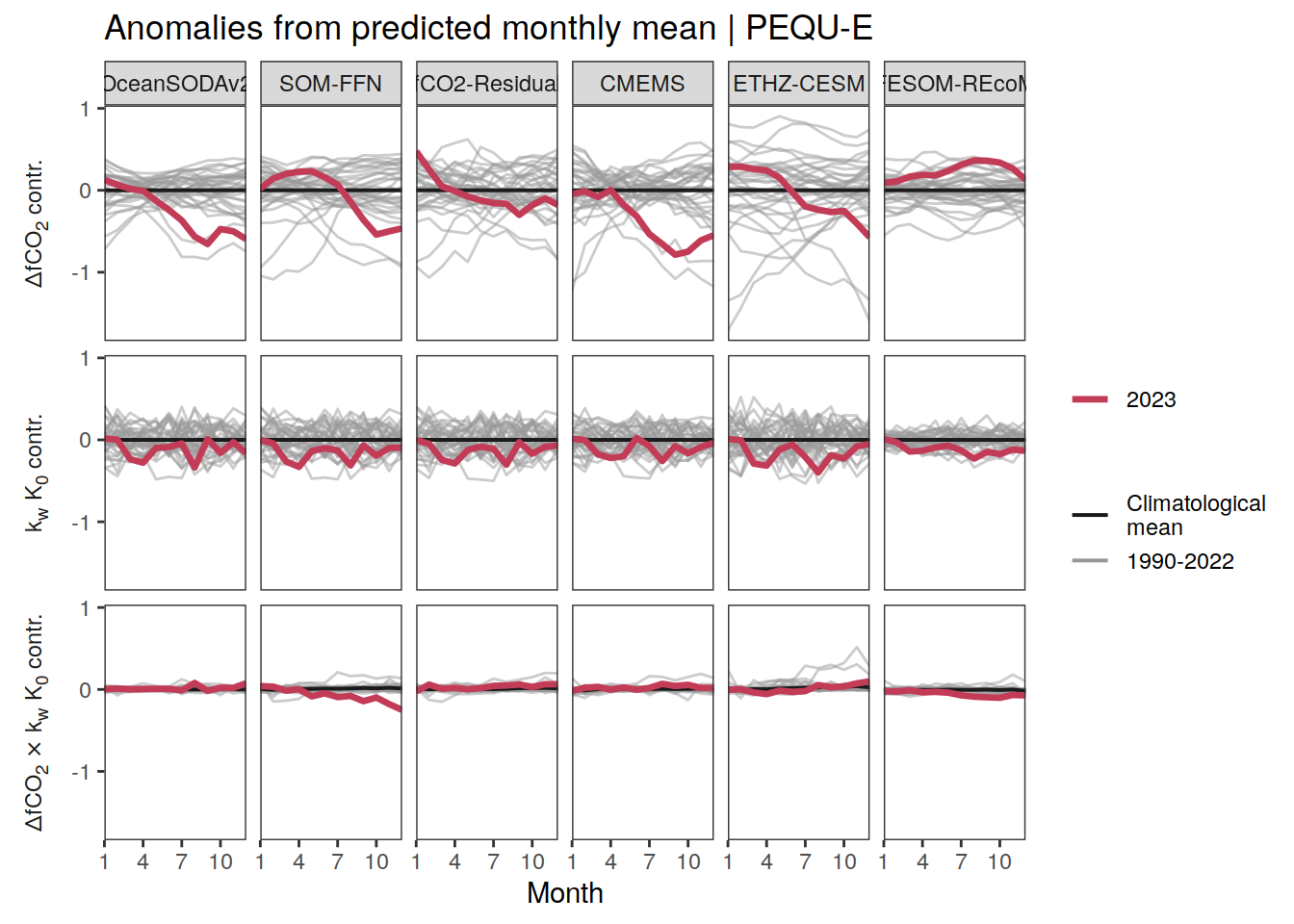
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
Annual
# pco2_product_biome_annual_flux_attribution <-
# full_join(
# pco2_product_biome_annual_flux_attribution %>%
# filter(year == 2023) %>%
# select(-year),
# pco2_product_biome_annual_flux_attribution %>%
# filter(year != 2023) %>%
# group_by(product, biome, name) %>%
# summarise(resid_mean = mean(abs(resid))) %>%
# ungroup())
pco2_product_biome_annual_flux_attribution %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_col(aes("x", resid, fill = product),
position = "dodge2") +
scale_fill_manual(values = color_products) +
geom_col(
aes(
"x",
resid_mean * sign(resid),
group = product,
col = paste0("Mean\nexcl.",2023)
),
position = "dodge2",
fill = "transparent"
) +
labs(y = labels_breaks(unique("fgco2"))$i_legend_title,
title = .x$biome) +
facet_grid(
.~name,
labeller = labeller(name = x_axis_labels),
switch = "x"
) +
scale_color_grey() +
theme(
legend.title = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_markdown(),
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
legend.position = "top"
)
)[[1]]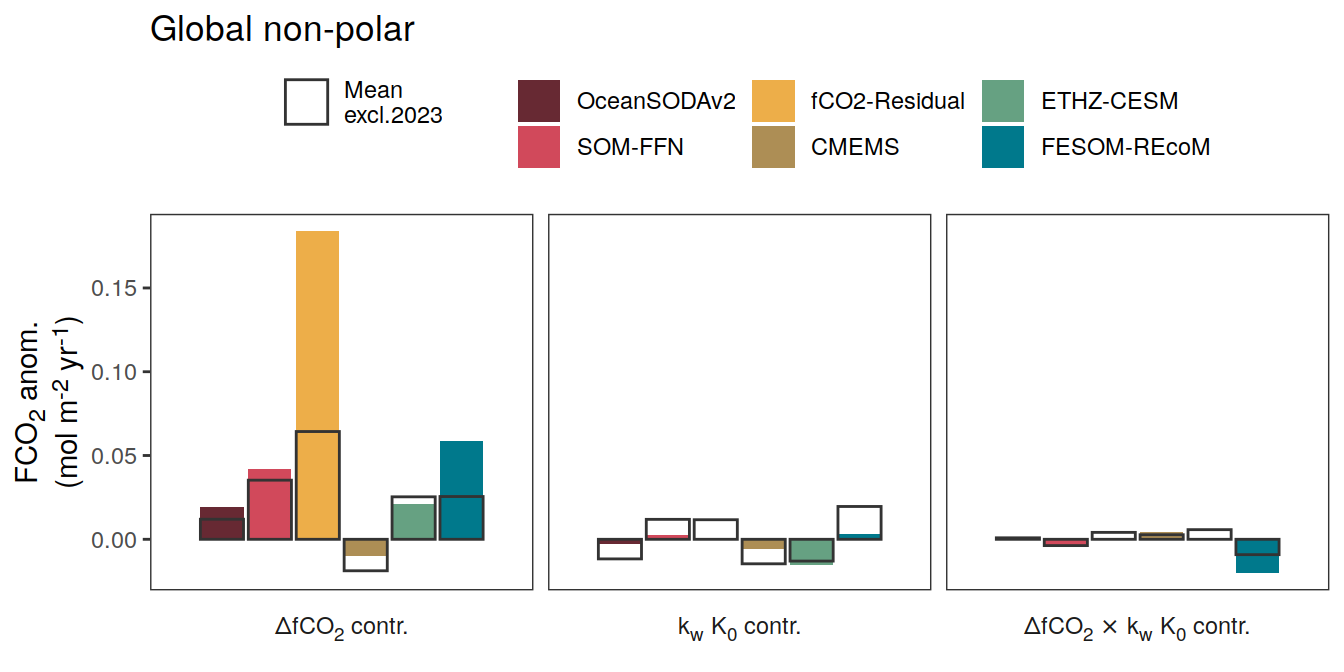
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
[[2]]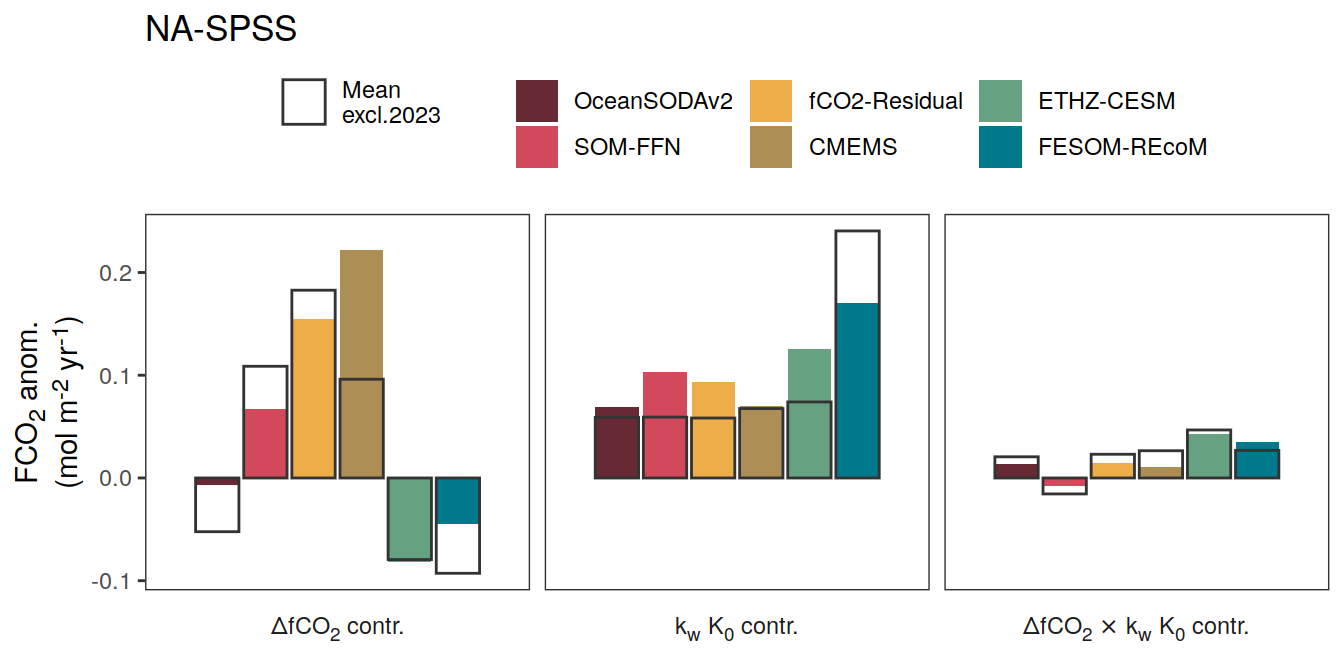
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
[[3]]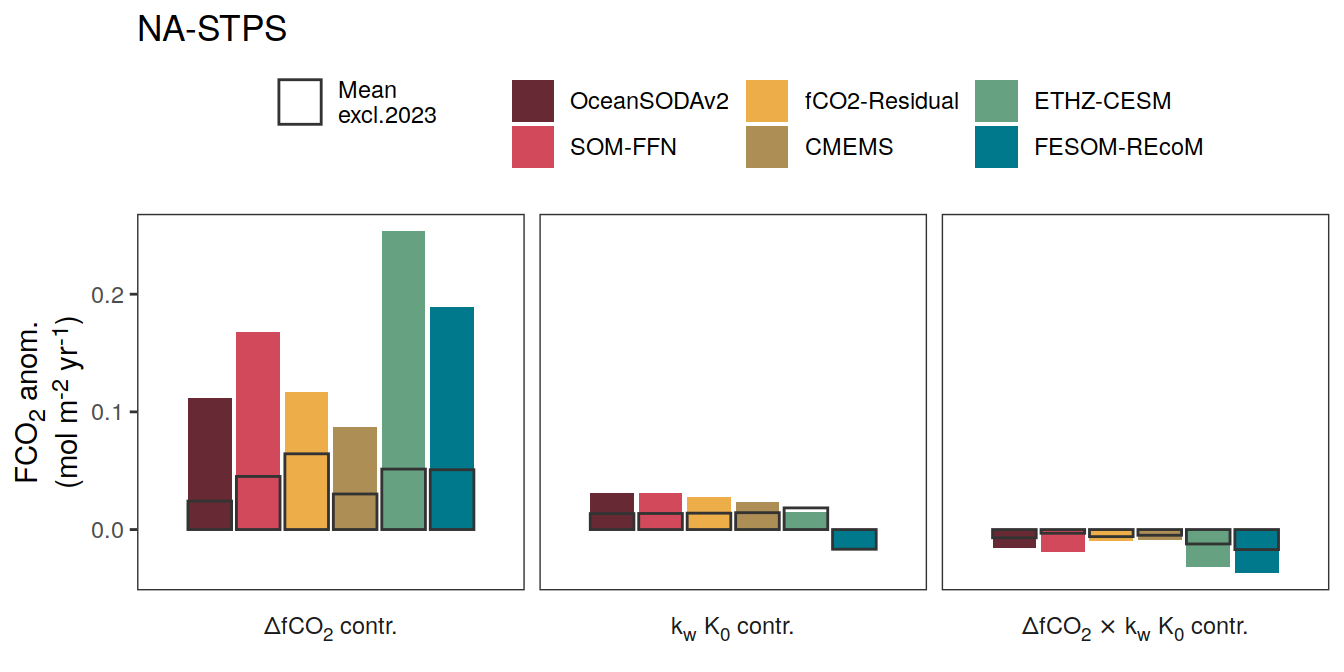
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
[[4]]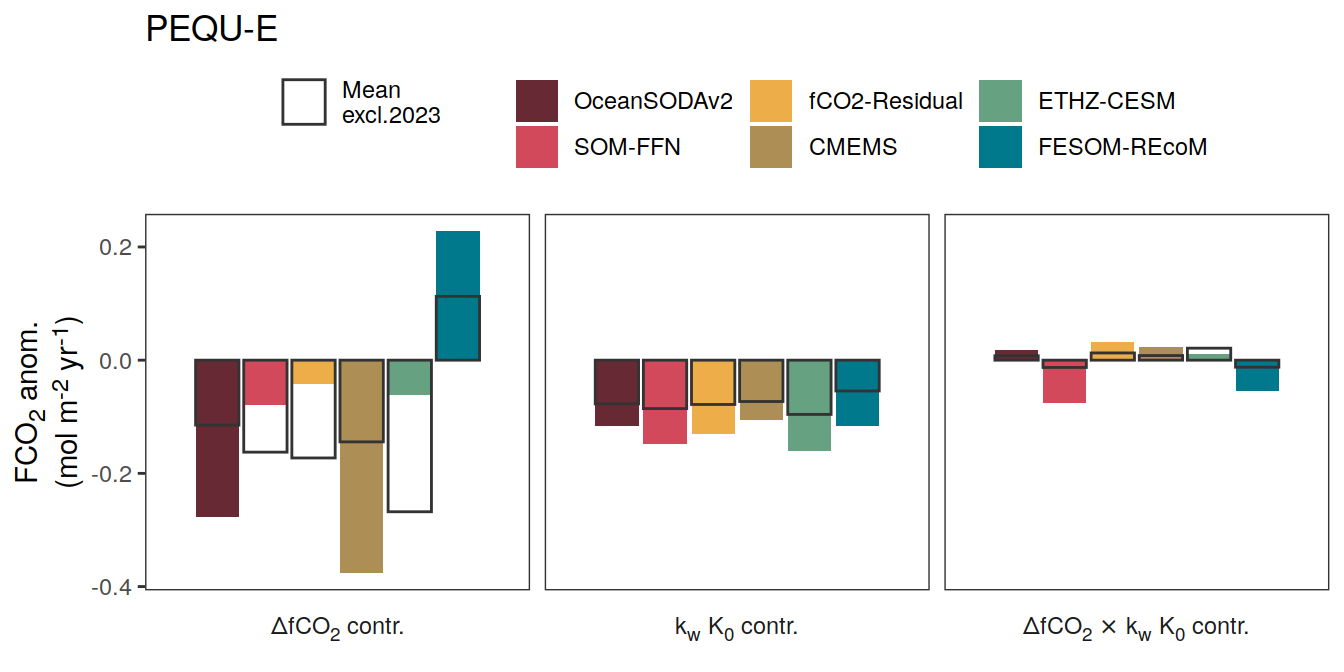
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| 7b6f27c | jens-daniel-mueller | 2024-05-27 |
| 29e0ec4 | jens-daniel-mueller | 2024-05-21 |
| 7c08e1c | jens-daniel-mueller | 2024-05-21 |
| dbc1fc6 | jens-daniel-mueller | 2024-05-16 |
| aea0b99 | jens-daniel-mueller | 2024-05-16 |
| 3310cf6 | jens-daniel-mueller | 2024-05-16 |
| b7d0689 | jens-daniel-mueller | 2024-05-15 |
Merged seasonality plots
pco2_product_biome_monthly_detrended %>%
filter(product %in% pco2_product_list) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup() %>%
filter(name %in% c("temperature", "fgco2"), biome %in% key_biomes,
year != 2023) %>%
group_by(month, biome, name) %>%
summarise(resid_sd = sd(resid)) %>%
ungroup() %>%
ggplot(aes(month, resid_sd)) +
geom_path() +
facet_grid(name ~ biome, scales = "free_y")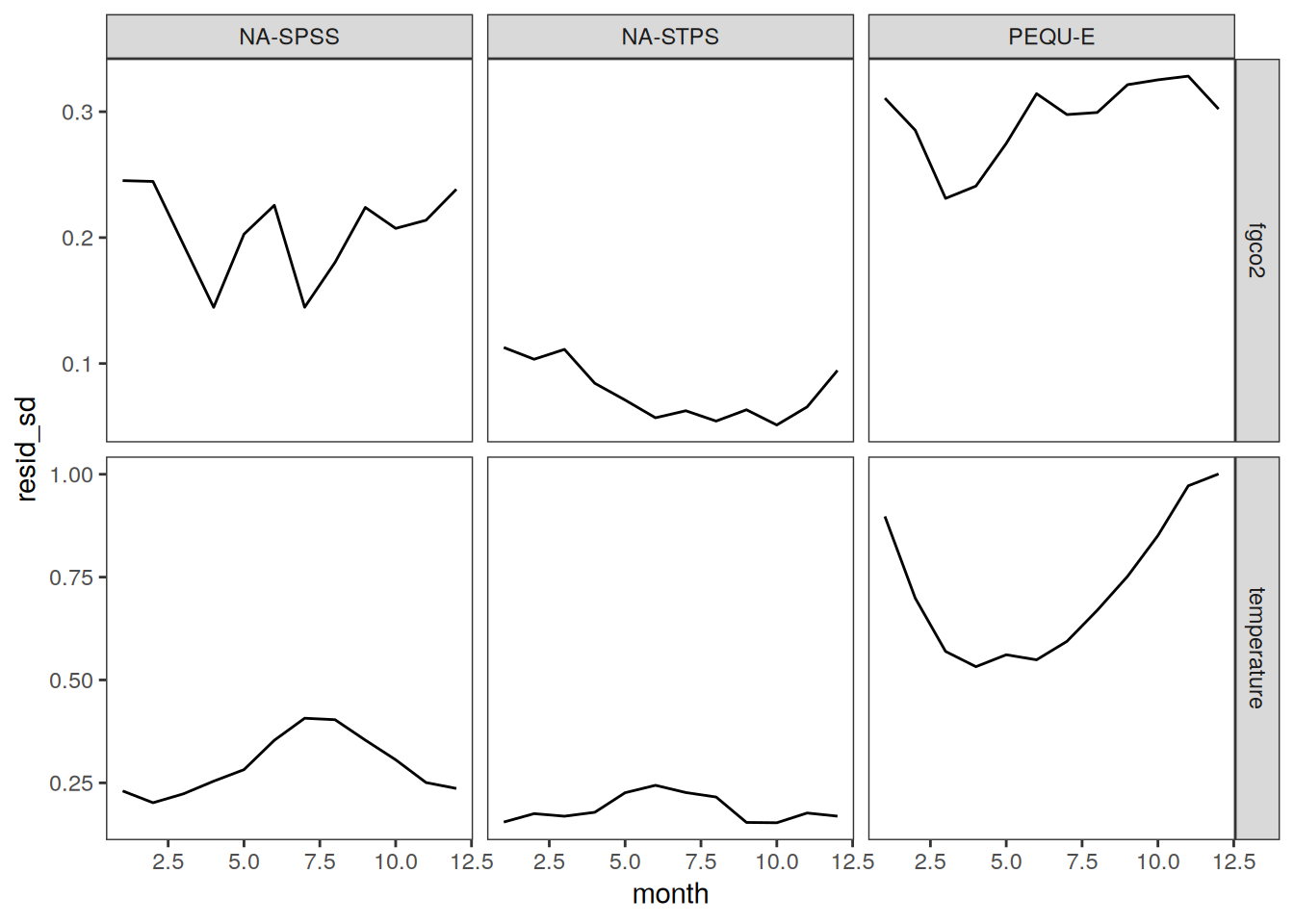
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| aeca619 | jens-daniel-mueller | 2024-06-19 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
pco2_product_biome_monthly_detrended %>%
filter(product %in% pco2_product_list) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup() %>%
filter(name %in% c("temperature", "fgco2"), biome %in% key_biomes) %>%
p_season(dim_col = "biome",
title = "Ensemble mean anomalies from predicted monthly mean") +
theme(axis.title.x = element_blank(), axis.text.x = element_blank()) +
new_scale_color() +
scale_color_manual(values = warm_cool_gradient[15]) +
geom_path(
data = pco2_product_biome_monthly_detrended %>%
filter(
product %in% gobm_product_list,
year == 2023,
name %in% c("temperature", "fgco2"),
biome %in% key_biomes
) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup(),
aes(month, resid, col = "2023\nGOBM mean")
)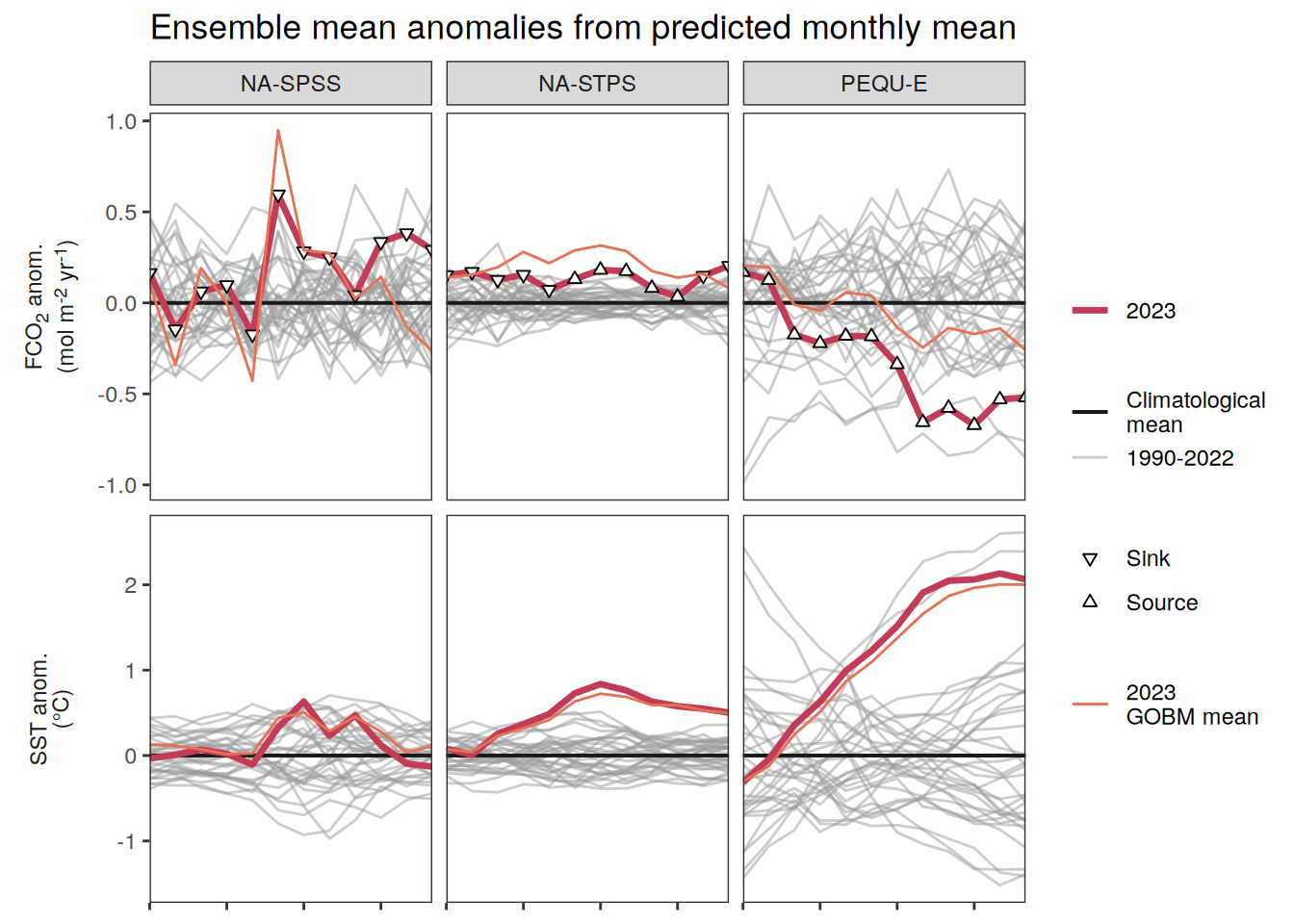
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| aeca619 | jens-daniel-mueller | 2024-06-19 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
ggsave(width = 9,
height = 4,
dpi = 600,
filename = "../output/biome_seasonal_anomaly_fgco2_sst_ensemble_mean_pco2_products.jpg")
pco2_product_biome_monthly_flux_attribution %>%
filter(product %in% pco2_product_list) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup() %>%
filter(name %in% c("resid_fgco2_dfco2", "resid_fgco2_kw_sol"),
biome %in% key_biomes) %>%
p_season(dim_col = "biome",
title = "Ensemble mean drivers of flux anomaly",
scales = "fixed") +
new_scale_color() +
scale_color_manual(values = warm_cool_gradient[15]) +
geom_path(
data = pco2_product_biome_monthly_flux_attribution %>%
filter(
product %in% gobm_product_list,
year == 2023,
name %in% c("resid_fgco2_dfco2", "resid_fgco2_kw_sol"),
biome %in% key_biomes
) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup(),
aes(month, resid, col = "2023\nGOBM mean")
)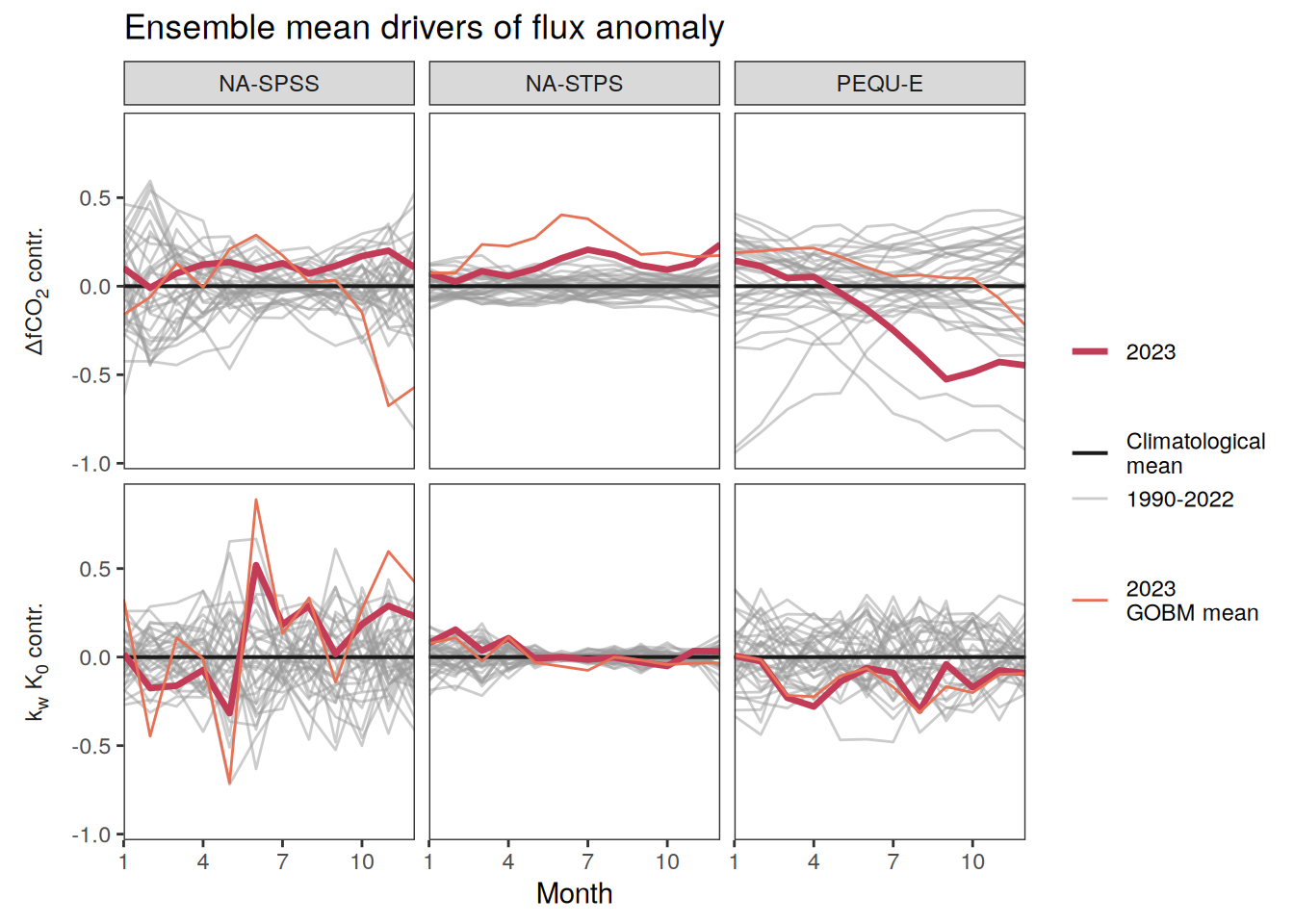
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| b18b0e5 | jens-daniel-mueller | 2024-06-28 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| aeca619 | jens-daniel-mueller | 2024-06-19 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| b754e95 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
ggsave(width = 9,
height = 4,
dpi = 600,
filename = "../output/biome_seasonal_anomaly_fgco2_attribution_ensemble_mean_pco2_products.jpg")
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(product %in% pco2_product_list) %>%
group_by(year, month, biome, name) %>%
summarise(across(where(is.numeric), mean)) %>%
ungroup() %>%
filter(name %in% c("sfco2_nontherm", "sfco2_therm", "sfco2_total"),
biome %in% c("Global non-polar", key_biomes)) %>%
p_season(dim_col = "biome",
title = "Ensemble mean decomposition of fCO2 anomaly") 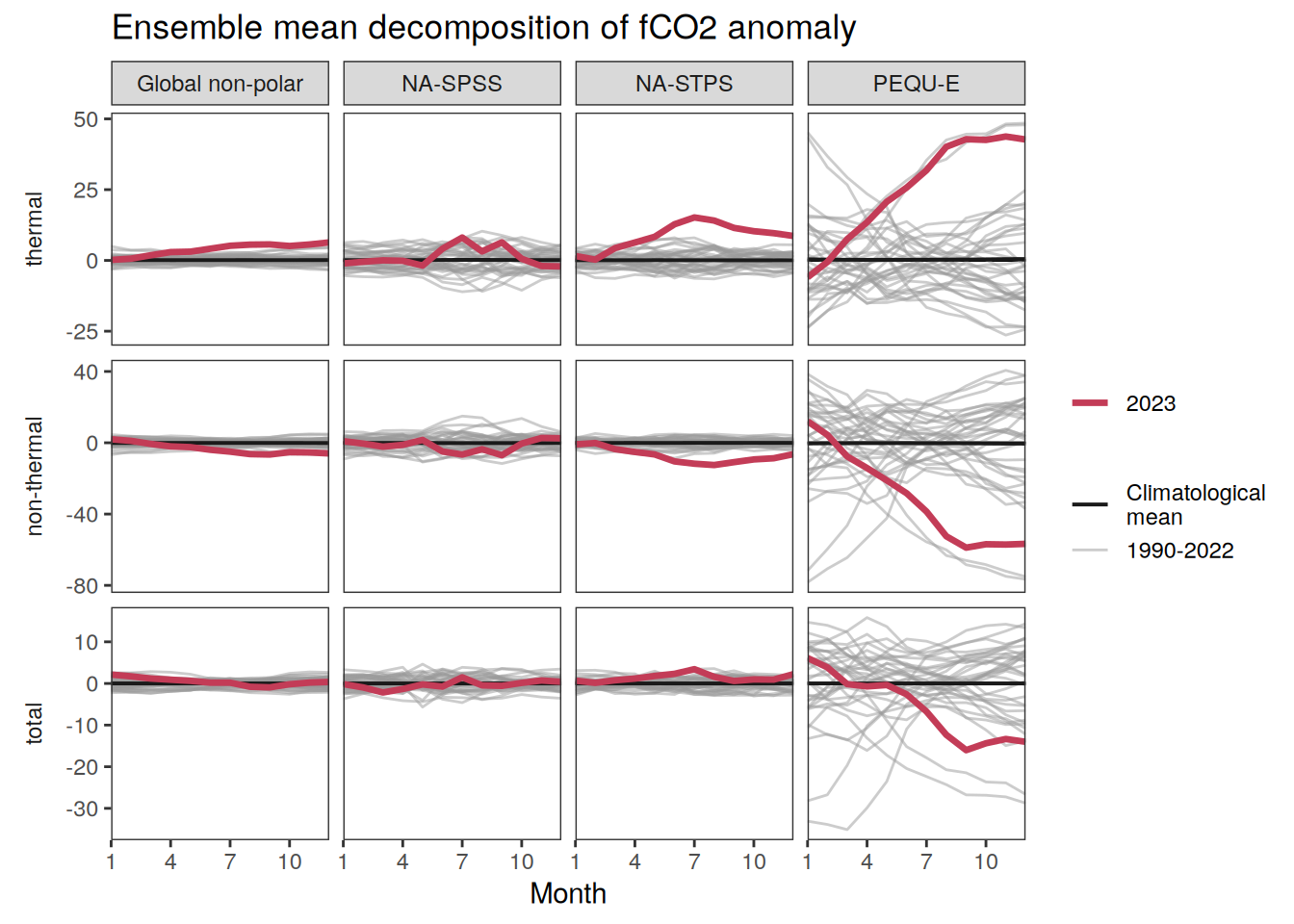
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| aecb187 | jens-daniel-mueller | 2024-08-28 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
ggsave(width = 9,
height = 4,
dpi = 600,
filename = "../output/biome_seasonal_anomaly_fco2_decomposition_ensemble_mean_pco2_products.jpg")pco2_product_biome_monthly_detrended %>%
filter(year == 2023,
name %in% c("temperature", "fgco2"),
biome %in% c("Global non-polar", key_biomes)) %>%
ggplot(aes(month, resid)) +
geom_hline(yintercept = 0, linewidth = 0.5) +
geom_path(aes(col = product)) +
scale_color_manual(values = color_products) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(x = "Month",
title = "Anomalies from predicted monthly mean") +
facet_grid(
name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)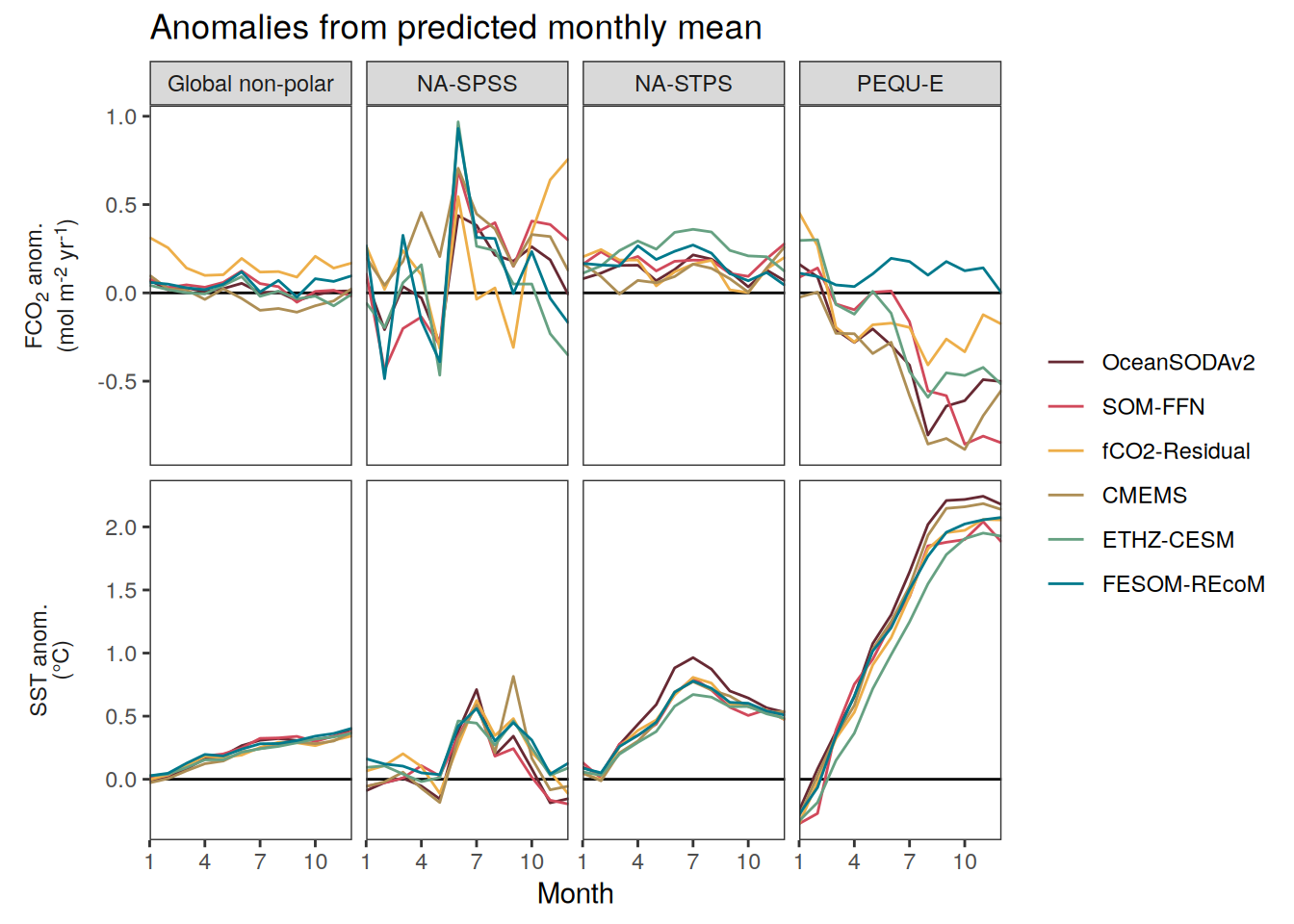
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| aeca619 | jens-daniel-mueller | 2024-06-19 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 7868a54 | jens-daniel-mueller | 2024-05-22 |
ggsave(width = 9,
height = 3,
dpi = 600,
filename = "../output/biome_seasonal_anomaly_fgco2_sst_all_products.jpg")
pco2_product_biome_monthly_flux_attribution %>%
filter(year == 2023,
name %in% c("resid_fgco2_dfco2", "resid_fgco2_kw_sol"),
biome %in% c("Global non-polar", key_biomes)) %>%
ggplot(aes(month, resid)) +
geom_hline(yintercept = 0, linewidth = 0.5) +
geom_path(aes(col = product)) +
scale_color_manual(values = color_products) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(x = "Month",
title = "Drivers of flux anomaly") +
facet_grid(
name ~ biome,
scales = "fixed",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)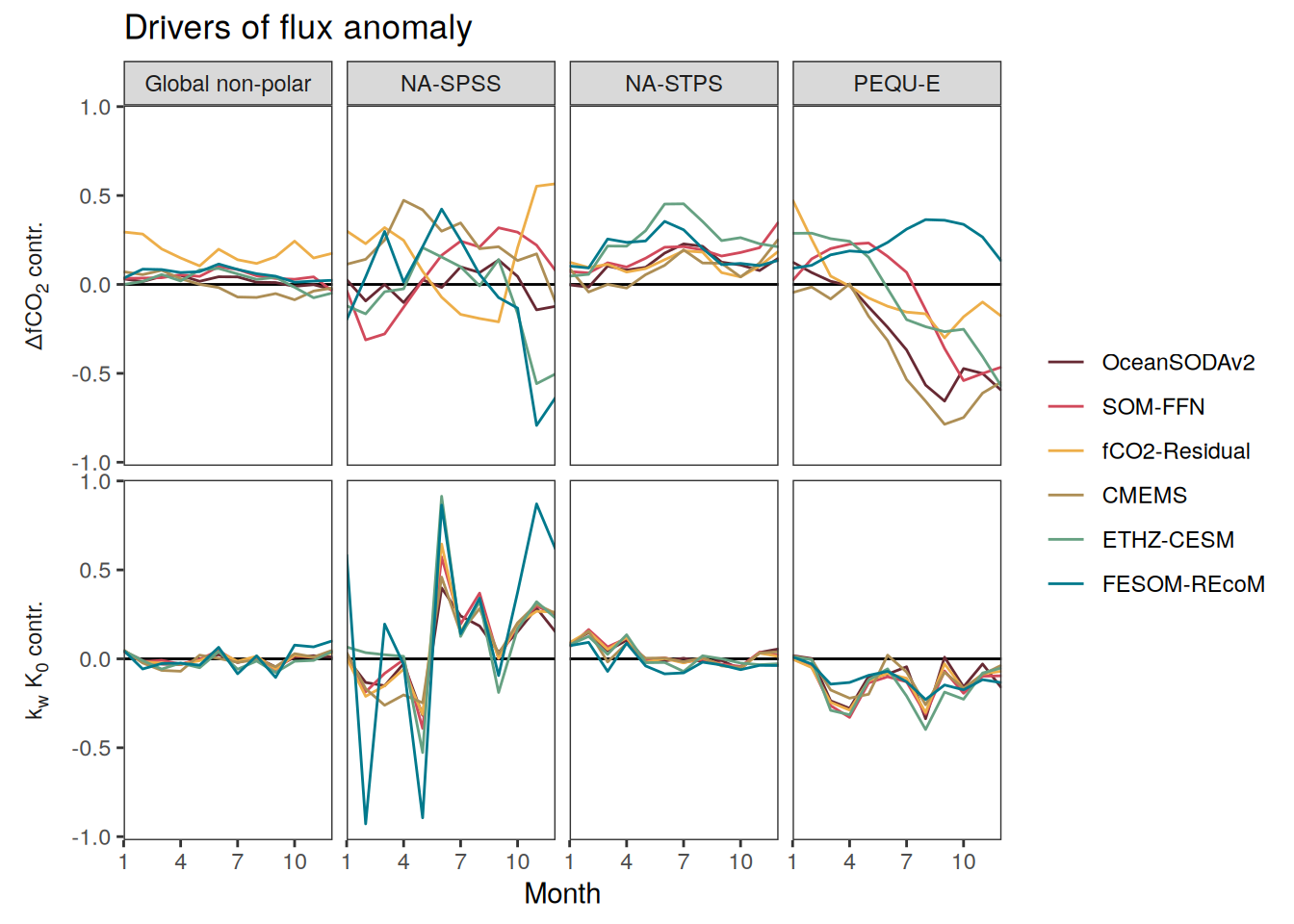
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 2f165ec | jens-daniel-mueller | 2024-07-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| aeca619 | jens-daniel-mueller | 2024-06-19 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 7868a54 | jens-daniel-mueller | 2024-05-22 |
ggsave(width = 9,
height = 3,
dpi = 600,
filename = "../output/biome_seasonal_anomaly_fgco2_attribution_all_products.jpg")
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(year == 2023,
name %in% c("sfco2_nontherm", "sfco2_therm", "sfco2_total"),
biome %in% c("Global non-polar", key_biomes)) %>%
ggplot(aes(month, resid)) +
geom_hline(yintercept = 0, linewidth = 0.5) +
geom_path(aes(col = product)) +
scale_color_manual(values = color_products) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(x = "Month",
title = "Decomposition of fCO2 anomaly") +
facet_grid(
name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)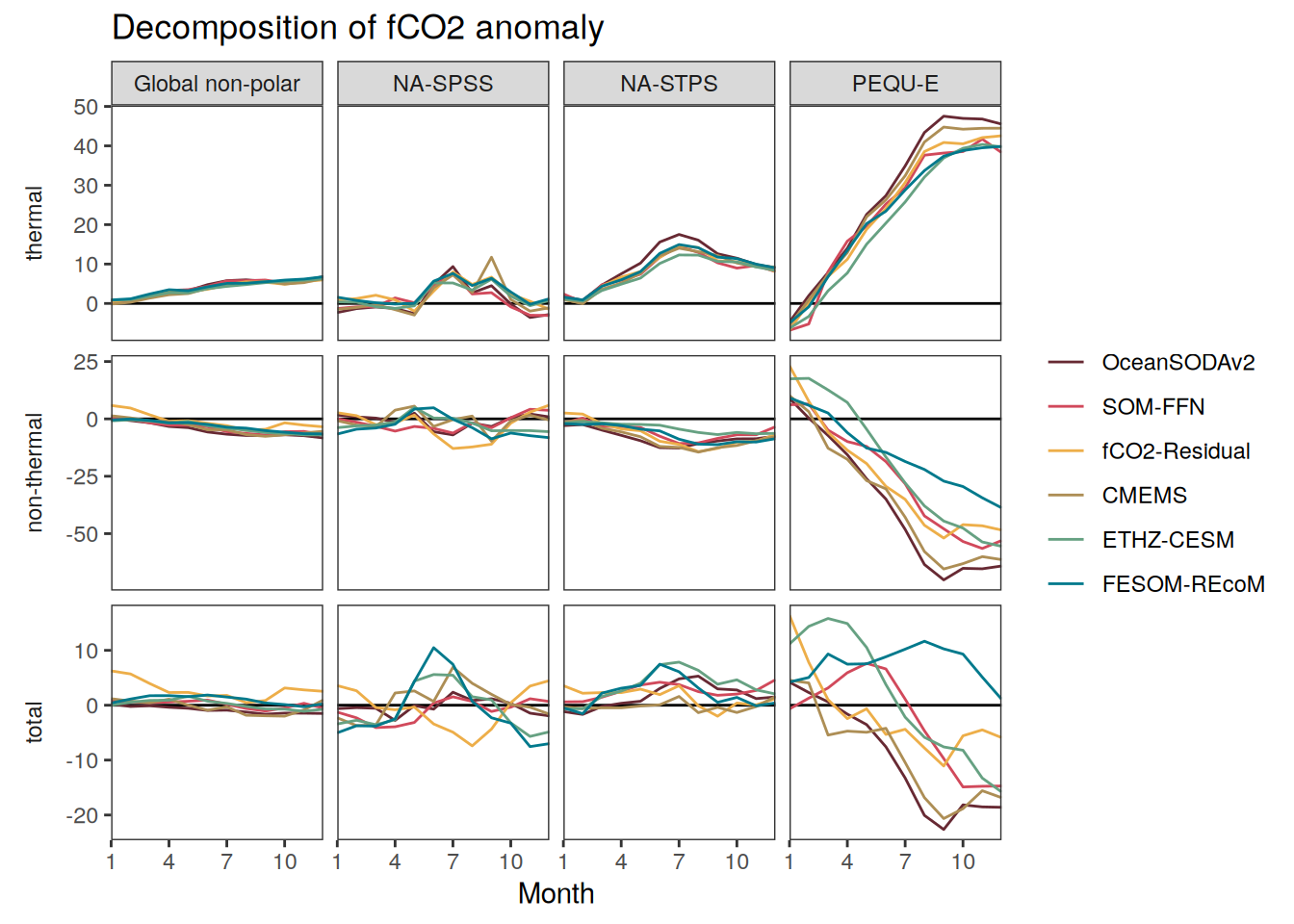
| Version | Author | Date |
|---|---|---|
| 709a2cf | jens-daniel-mueller | 2025-02-26 |
| 4d090e2 | jens-daniel-mueller | 2025-02-25 |
| 3548f48 | jens-daniel-mueller | 2025-02-25 |
| 1d3f08c | jens-daniel-mueller | 2025-02-24 |
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| d3235fa | jens-daniel-mueller | 2024-09-20 |
| 878c674 | jens-daniel-mueller | 2024-09-10 |
| c62d92d | jens-daniel-mueller | 2024-08-23 |
| 4ef742b | jens-daniel-mueller | 2024-07-20 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| 4a437fb | jens-daniel-mueller | 2024-07-09 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 67956dd | jens-daniel-mueller | 2024-07-08 |
| b7806ad | jens-daniel-mueller | 2024-07-02 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 9589349 | jens-daniel-mueller | 2024-06-27 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| aeca619 | jens-daniel-mueller | 2024-06-19 |
| d2a80a9 | jens-daniel-mueller | 2024-06-14 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 9cfceb9 | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| b99b329 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| fe97ed3 | jens-daniel-mueller | 2024-05-25 |
| 7868a54 | jens-daniel-mueller | 2024-05-22 |
ggsave(width = 9,
height = 4,
dpi = 600,
filename = "../output/biome_seasonal_anomaly_fco2_decomposition_all_products.jpg")Biome profiles
The following analysis is available for GOBMs only.
Annual means
2023 anomaly
pco2_product_profiles_annual %>%
filter(biome %in% key_biomes,
name %in% name_core) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_vline(xintercept = 0) +
geom_path(aes(resid, depth, group = year), col = "grey30", alpha = 0.3) +
geom_path(data = .x %>% filter(year == 2023),
aes(resid, depth, col = as.factor(year)),
linewidth = 1) +
scale_color_brewer(palette = "Set1") +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50,100,200,400)) +
coord_cartesian(expand = 0) +
facet_grid2(biome ~ product,
scales = "free_x", independent = "x") +
labs(y = "Depth (m)",
x = labels_breaks(.x %>% distinct(name))$i_legend_title) +
theme(legend.title = element_blank(),
axis.title.x = element_markdown())
)[[1]]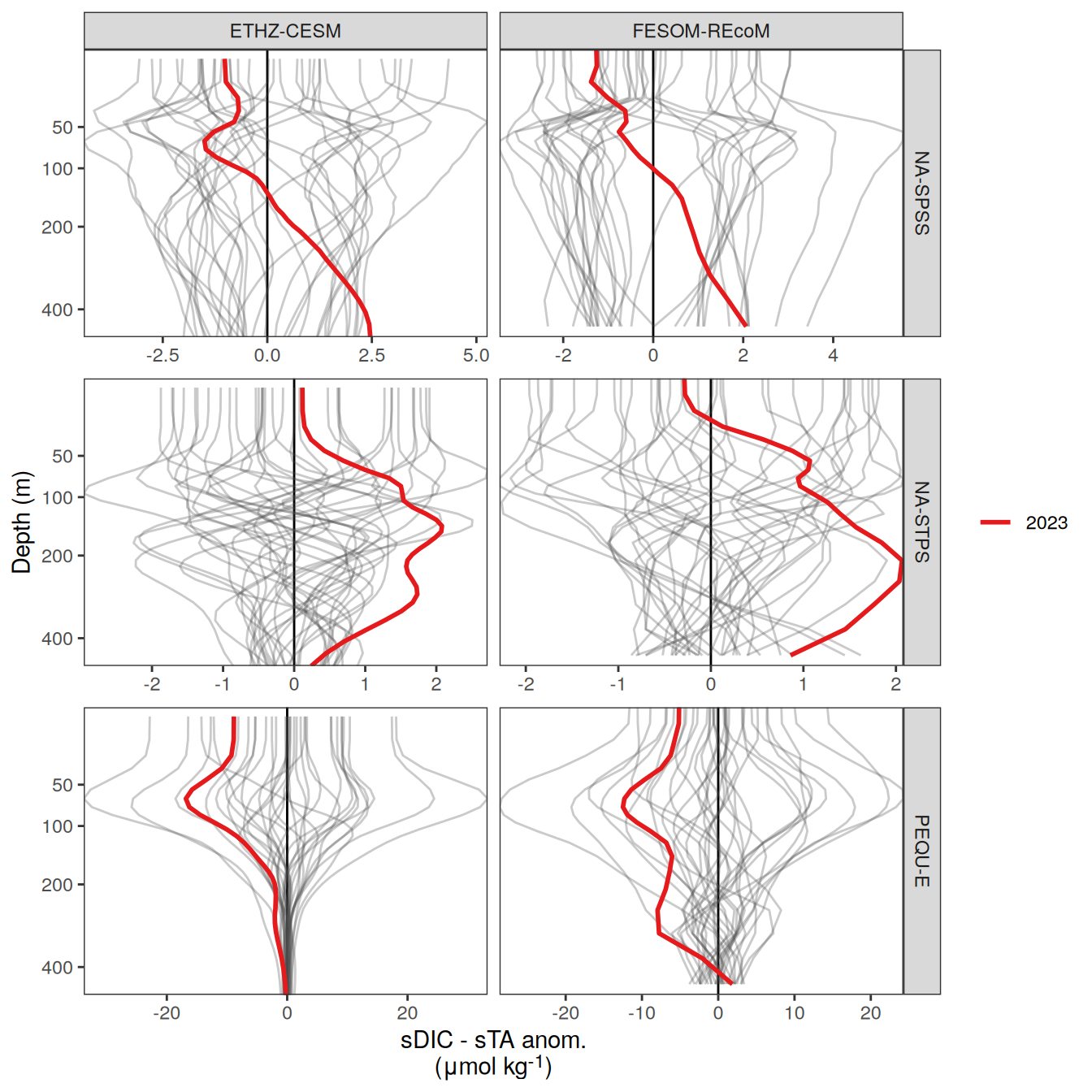
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
[[2]]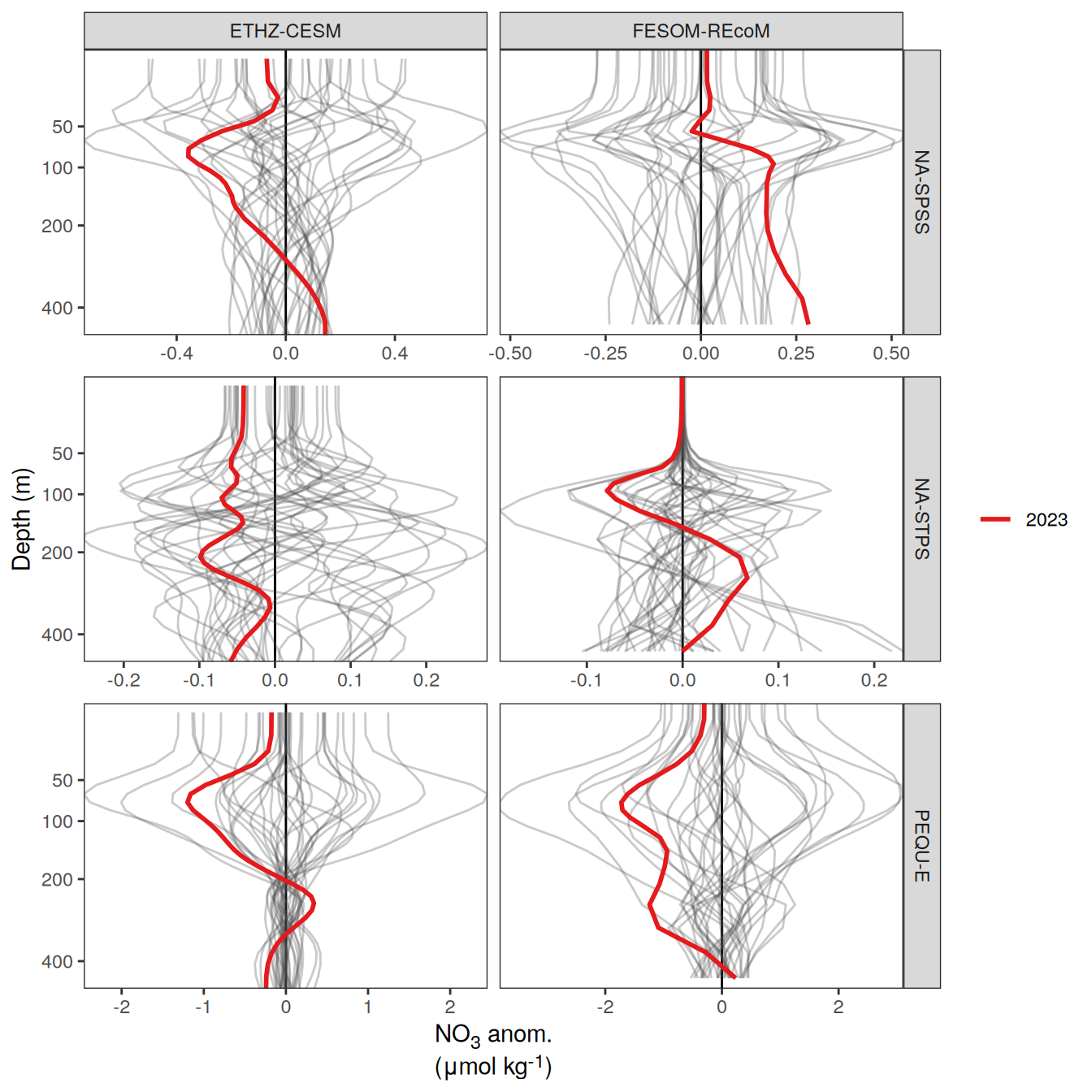
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
[[3]]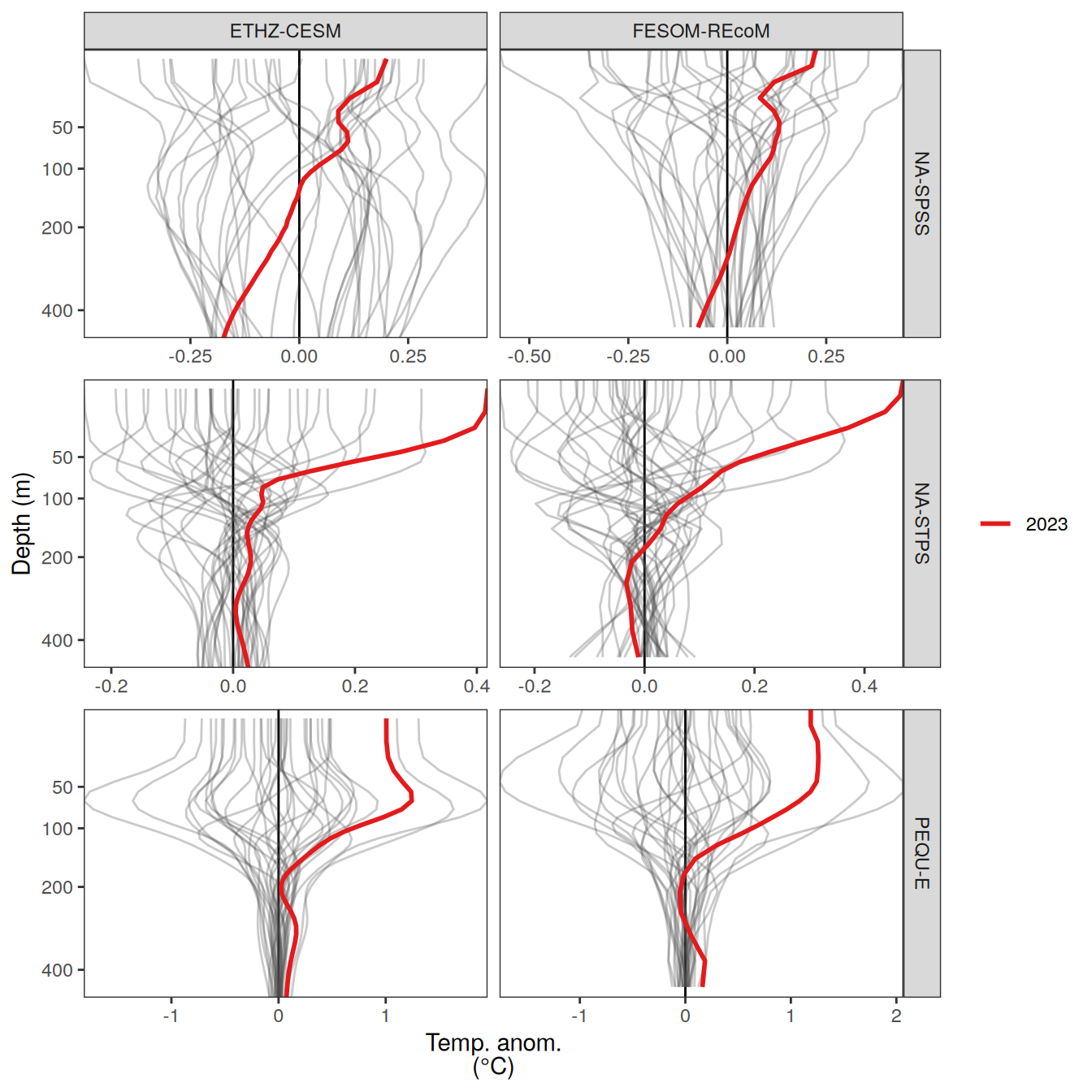
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
Surface anomaly conversion
pco2_product_profiles_monthly_conversion <-
read_csv(c("../data/ETHZ-CESM_2023_profiles_monthly.csv",
"../data/FESOM-REcoM_2023_profiles_monthly.csv"),
id = "product")
library(seacarb)
pco2_product_profiles_monthly_conversion <-
pco2_product_profiles_monthly_conversion %>%
filter(biome %in% key_biomes,
name %in% c("stalk", "sdissic", "thetao"),
year == 2023,
depth <= 5) %>%
select(product, name, biome, month, value, resid) %>%
group_by(name, biome, month) %>%
summarise(value = mean(value),
resid = mean(resid)) %>%
ungroup() %>%
pivot_wider(values_from = c(value, resid)) %>%
mutate(buffesm(
flag = 15,
var1 = value_stalk * 1e-6,
var2 = value_sdissic * 1e-6,
T = value_thetao
)[c(1, 4)],
carb(
flag = 15,
var1 = value_stalk * 1e-6,
var2 = value_sdissic * 1e-6,
T = value_thetao
)[c(8)]) %>%
mutate(
resid_fco2_stalk = gammaALK^-1 * fCO2 * resid_stalk * 1e-6,
resid_fco2_sdissic = gammaALK^-1 * fCO2 * resid_sdissic * 1e-6,
resid_fco2 = resid_fco2_stalk + resid_fco2_sdissic
) %>%
select(biome, month, starts_with("resid")) %>%
pivot_longer(starts_with("resid")) %>%
arrange(month) %>%
group_by(biome, name) %>%
mutate(value = value - first(value)) %>%
ungroup()
pco2_product_profiles_monthly_conversion %>%
ggplot(aes(month, value)) +
geom_hline(yintercept = 0) +
geom_path() +
facet_grid(biome ~ name, scales = "free_y")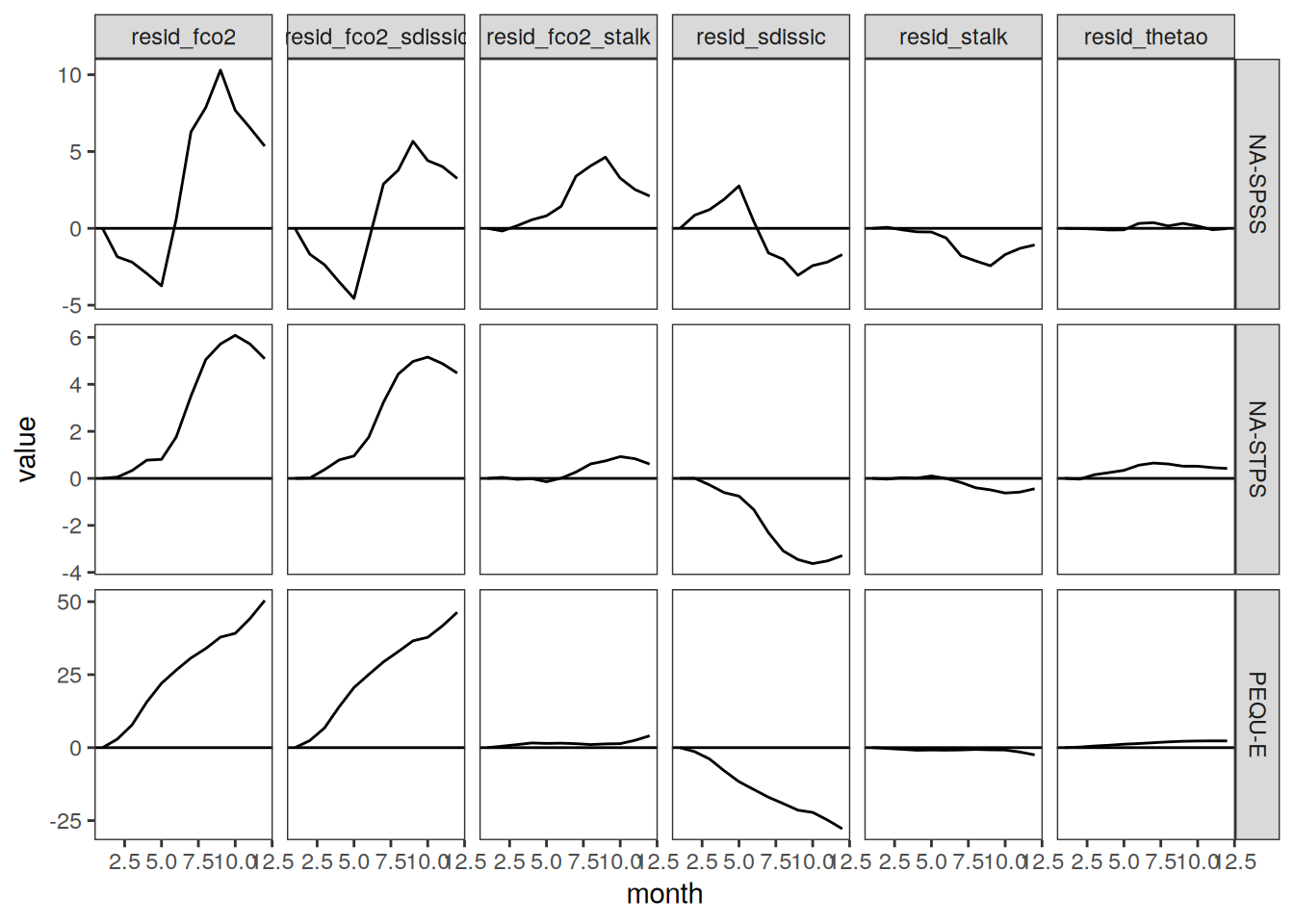
| Version | Author | Date |
|---|---|---|
| 3a5d484 | jens-daniel-mueller | 2025-03-11 |
pco2_product_profiles_monthly_conversion %>%
filter(month == 12)# A tibble: 18 × 4
biome month name value
<chr> <dbl> <chr> <dbl>
1 NA-SPSS 12 resid_sdissic -1.71
2 NA-SPSS 12 resid_stalk -1.08
3 NA-SPSS 12 resid_thetao -0.0169
4 NA-SPSS 12 resid_fco2_stalk 2.10
5 NA-SPSS 12 resid_fco2_sdissic 3.25
6 NA-SPSS 12 resid_fco2 5.35
7 NA-STPS 12 resid_sdissic -3.29
8 NA-STPS 12 resid_stalk -0.441
9 NA-STPS 12 resid_thetao 0.422
10 NA-STPS 12 resid_fco2_stalk 0.610
11 NA-STPS 12 resid_fco2_sdissic 4.48
12 NA-STPS 12 resid_fco2 5.09
13 PEQU-E 12 resid_sdissic -27.8
14 PEQU-E 12 resid_stalk -2.48
15 PEQU-E 12 resid_thetao 2.31
16 PEQU-E 12 resid_fco2_stalk 4.06
17 PEQU-E 12 resid_fco2_sdissic 46.3
18 PEQU-E 12 resid_fco2 50.4 Monthly means
2023 anomaly
pco2_product_profiles_monthly %>%
filter(year == 2023,
biome %in% key_biomes,
name %in% name_core) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_vline(xintercept = 0) +
geom_path(aes(resid, depth, col = as.factor(month)),
linewidth = 1) +
scale_color_viridis_d(option = "magma", end = .8) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50,100,200,400)) +
coord_cartesian(expand = 0) +
facet_grid2(biome ~ product,
scales = "free_x", independent = "x") +
labs(y = "Depth (m)",
x = labels_breaks(.x %>% distinct(name))$i_legend_title) +
theme(legend.title = element_blank(),
axis.title.x = element_markdown())
)[[1]]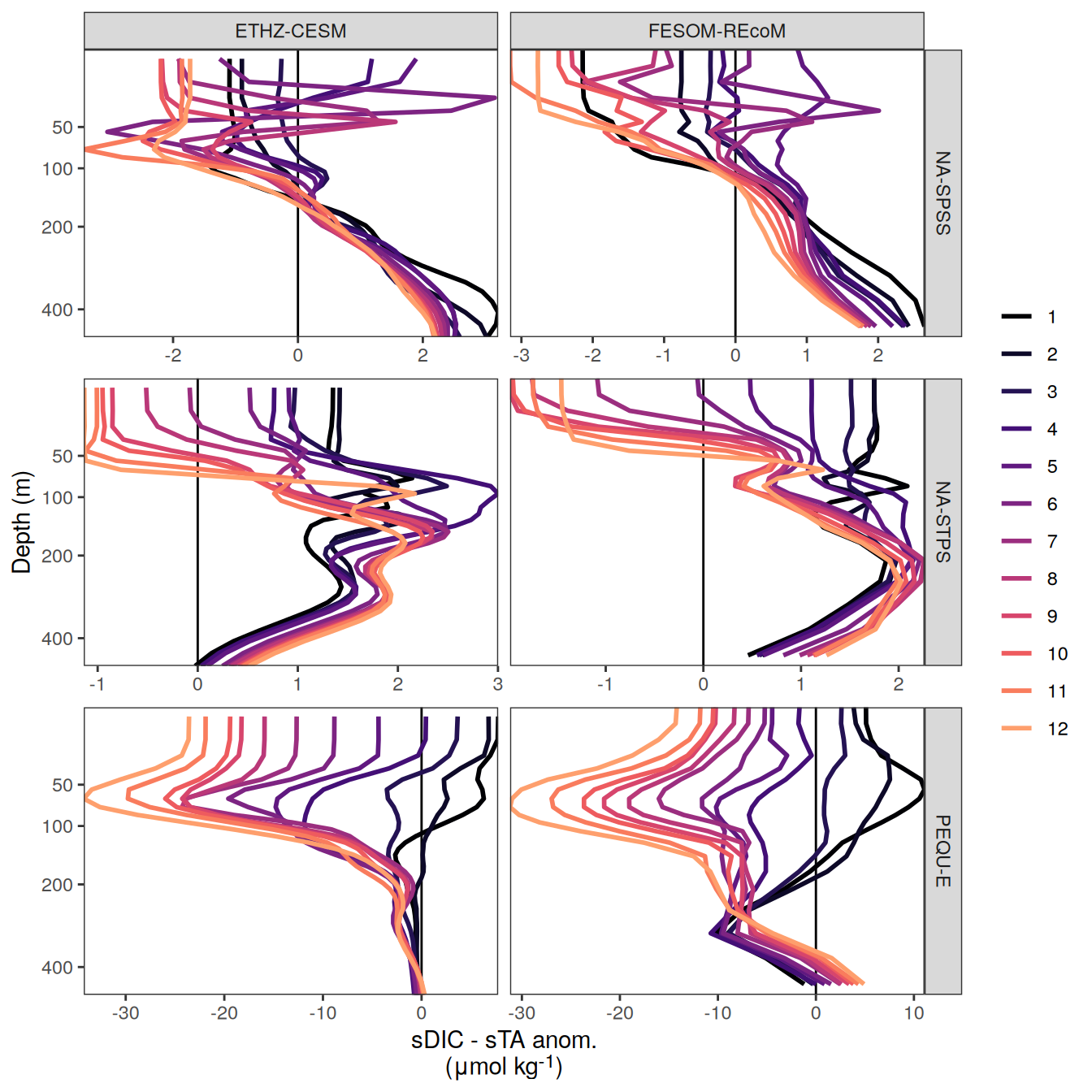
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
[[2]]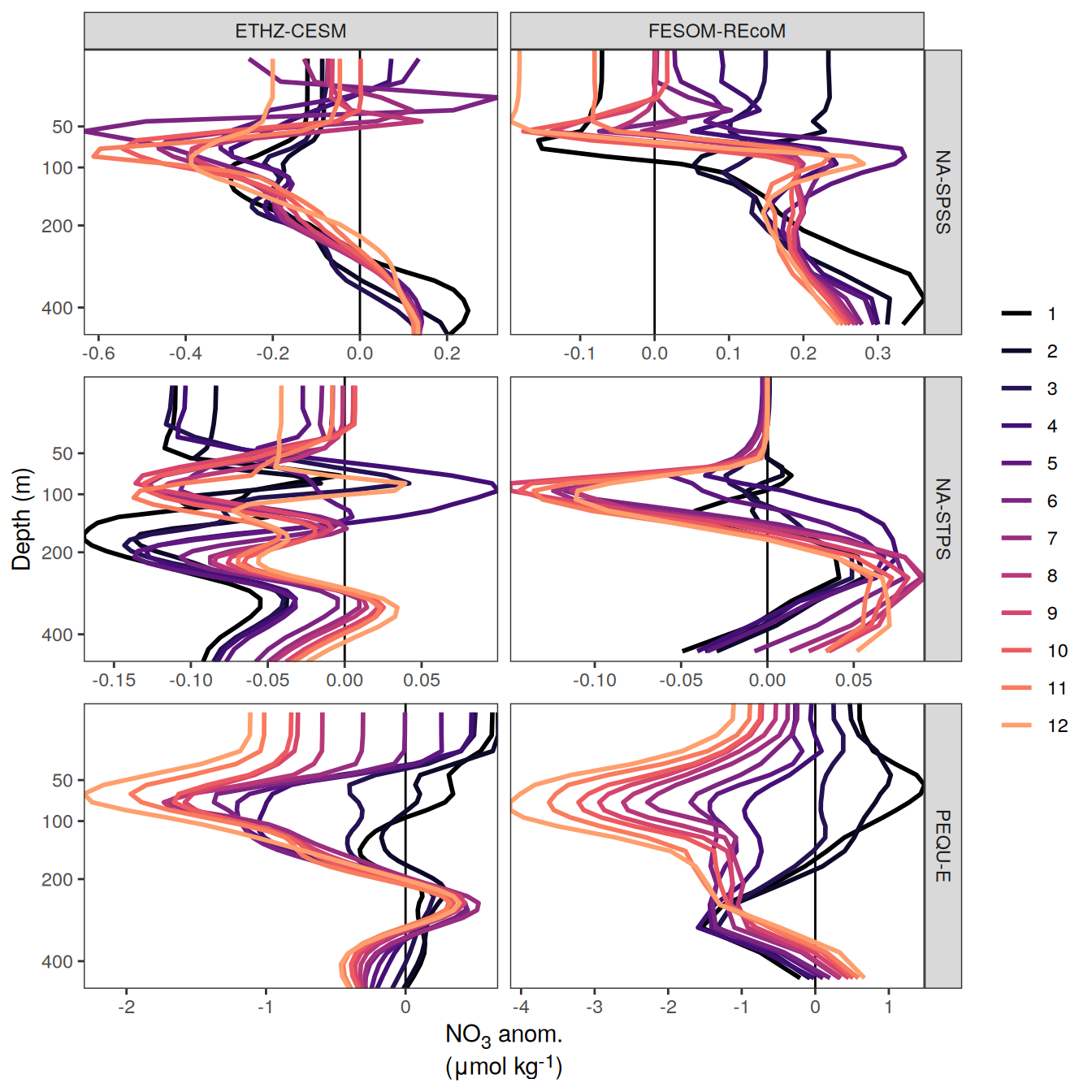
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
[[3]]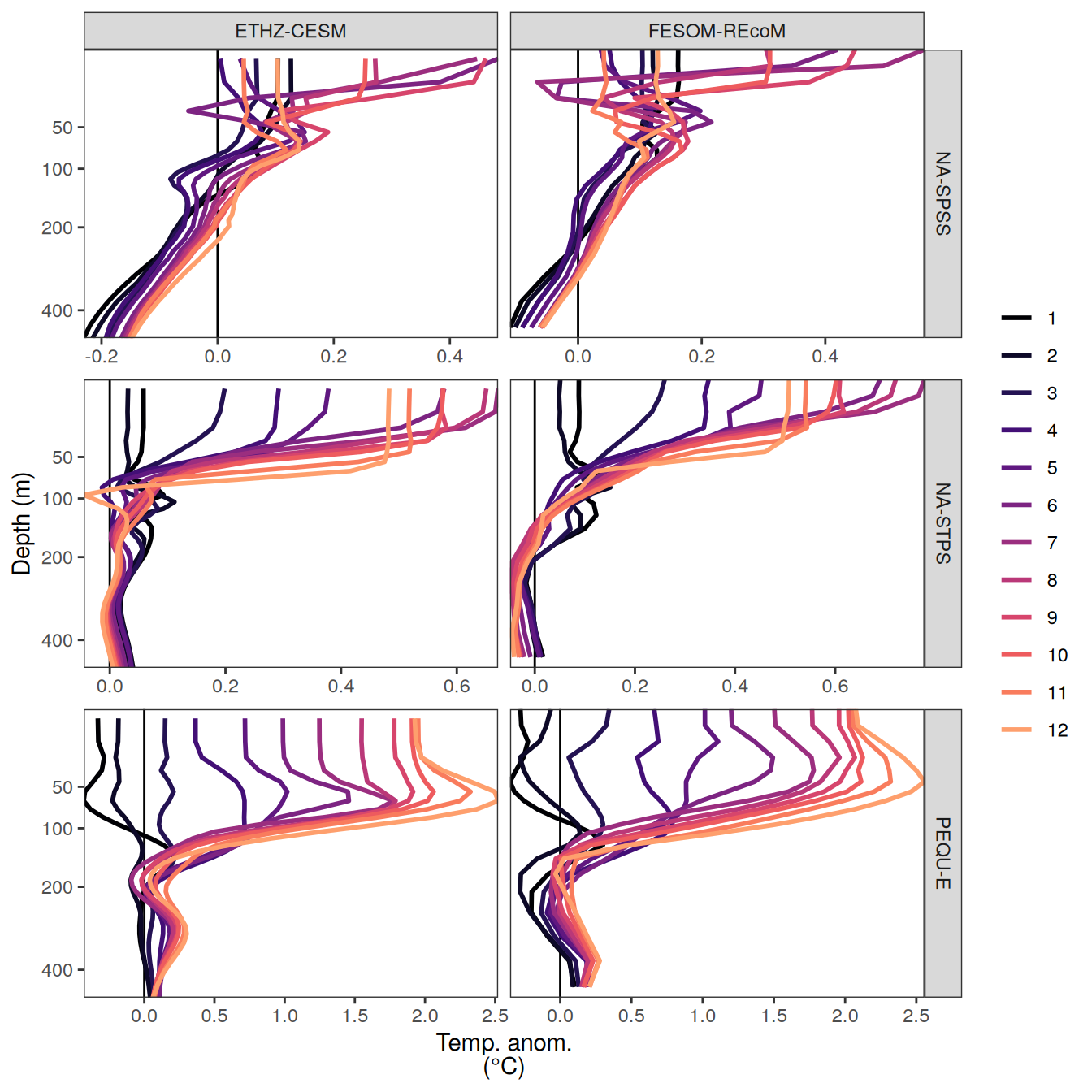
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| a58162a | jens-daniel-mueller | 2024-07-11 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| fbba0a0 | jens-daniel-mueller | 2024-05-28 |
| d533f68 | jens-daniel-mueller | 2024-05-28 |
| 7013182 | jens-daniel-mueller | 2024-05-27 |
| 97eff6a | jens-daniel-mueller | 2024-05-25 |
pco2_product_profiles_monthly %>%
filter(year == 2023,
biome %in% key_biomes,
product == "ETHZ-CESM",
name %in% name_core) %>%
ggplot() +
geom_vline(xintercept = 0) +
geom_path(aes(resid, depth, col = as.factor(month)),
linewidth = 1) +
scale_color_viridis_d(option = "magma", end = .8,
name = paste("Month of\n", 2023)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50, 100, 200, 400)) +
coord_cartesian(expand = 0) +
facet_grid2(
biome ~ name,
scales = "free_x",
independent = "x",
labeller = labeller(name = x_axis_labels),
switch = "x"
) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.x = element_blank()
) +
labs(y = "Depth (m)",
title = "Anomalies from monthly baseline (deseasonalized)")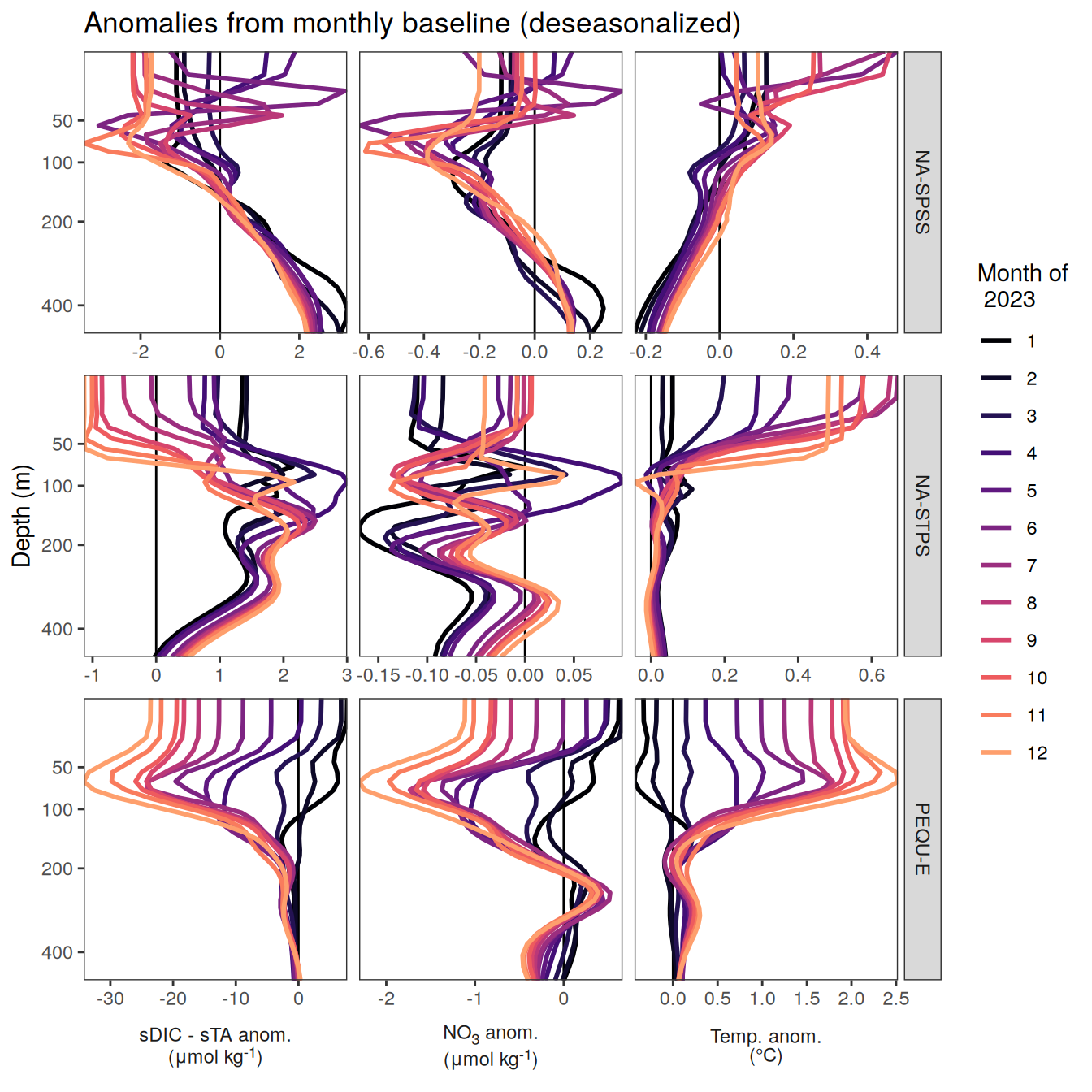
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| 8cdfed7 | jens-daniel-mueller | 2024-06-21 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| e83b65a | jens-daniel-mueller | 2024-05-31 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| fc1b92d | jens-daniel-mueller | 2024-05-30 |
| 4d3ccb2 | jens-daniel-mueller | 2024-05-29 |
| acaac5f | jens-daniel-mueller | 2024-05-28 |
# ggsave(width = 10,
# height = 8,
# dpi = 600,
# filename = "../output/CESM_2023_anomaly_profiles.jpg")
pco2_product_profiles_monthly %>%
filter(year == 2023,
biome %in% key_biomes,
product == "ETHZ-CESM",
name %in% name_core) %>%
arrange(month) %>%
group_by(biome, name, depth) %>%
mutate(resid = resid - first(resid)) %>%
ungroup() %>%
ggplot() +
geom_vline(xintercept = 0) +
geom_path(aes(resid, depth, col = as.factor(month)),
linewidth = 1) +
scale_color_viridis_d(option = "magma", end = .8,
name = paste("Month of\n", 2023)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50, 100, 200, 400)) +
coord_cartesian(expand = 0) +
facet_grid2(
biome ~ name,
scales = "free_x",
independent = "x",
labeller = labeller(name = x_axis_labels),
switch = "x"
) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.x = element_blank()
) +
labs(y = "Depth (m)",
title = "Monthly anomaly evolution relative to January 2023")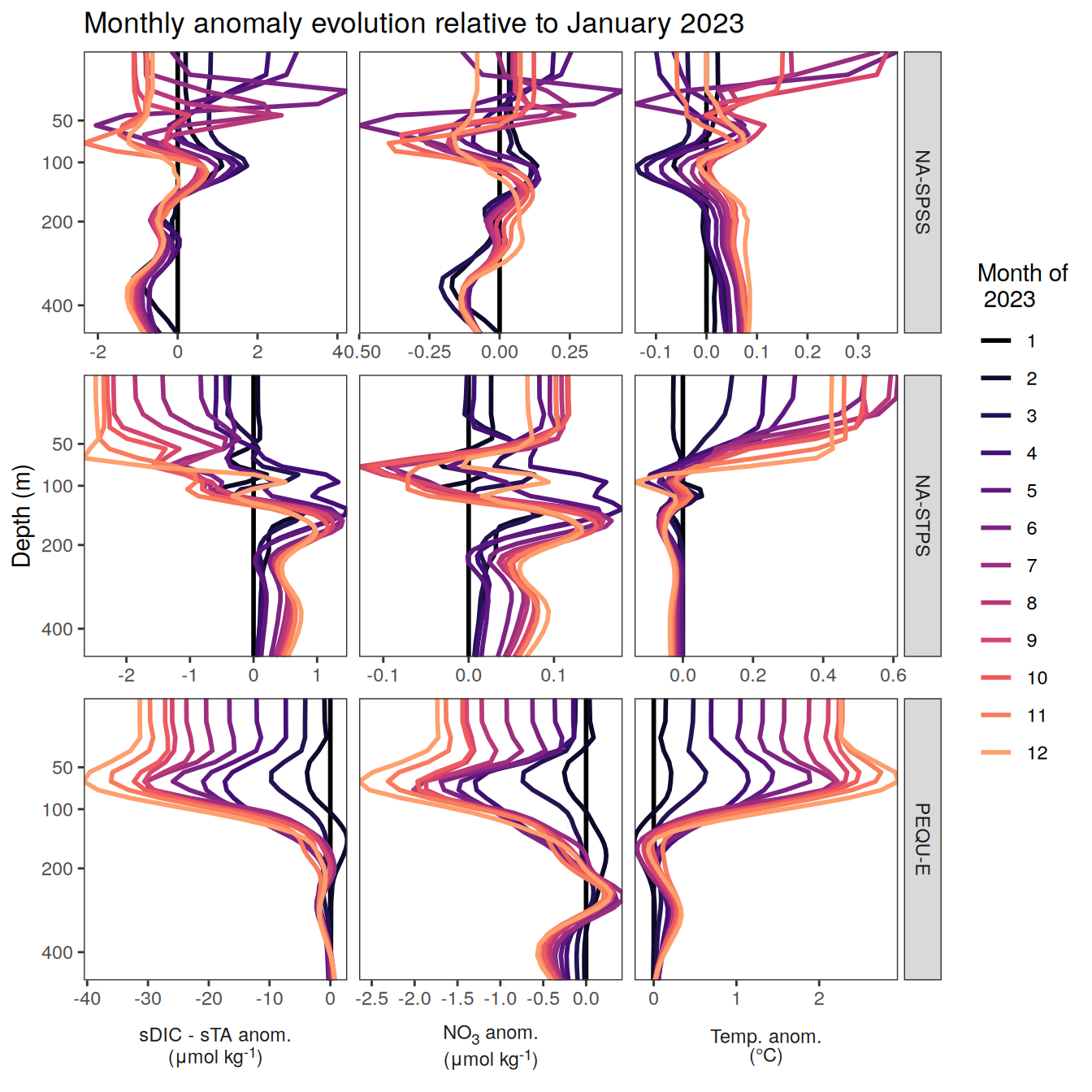
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| 2a28b07 | jens-daniel-mueller | 2024-07-22 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 6fc213f | jens-daniel-mueller | 2024-05-31 |
| fc1b92d | jens-daniel-mueller | 2024-05-30 |
pco2_product_profiles_monthly %>%
filter(year == 2023,
biome %in% key_biomes,
product == "FESOM-REcoM",
name %in% name_core) %>%
ggplot() +
geom_vline(xintercept = 0) +
geom_path(aes(resid, depth, col = as.factor(month)),
linewidth = 1) +
scale_color_viridis_d(option = "magma", end = .8,
name = paste("Month of\n", 2023)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50, 100, 200, 400)) +
coord_cartesian(expand = 0) +
facet_grid2(
biome ~ name,
scales = "free_x",
independent = "x",
labeller = labeller(name = x_axis_labels),
switch = "x"
) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.x = element_blank()
) +
labs(y = "Depth (m)",
title = "Anomalies from monthly baseline (deseasonalized)")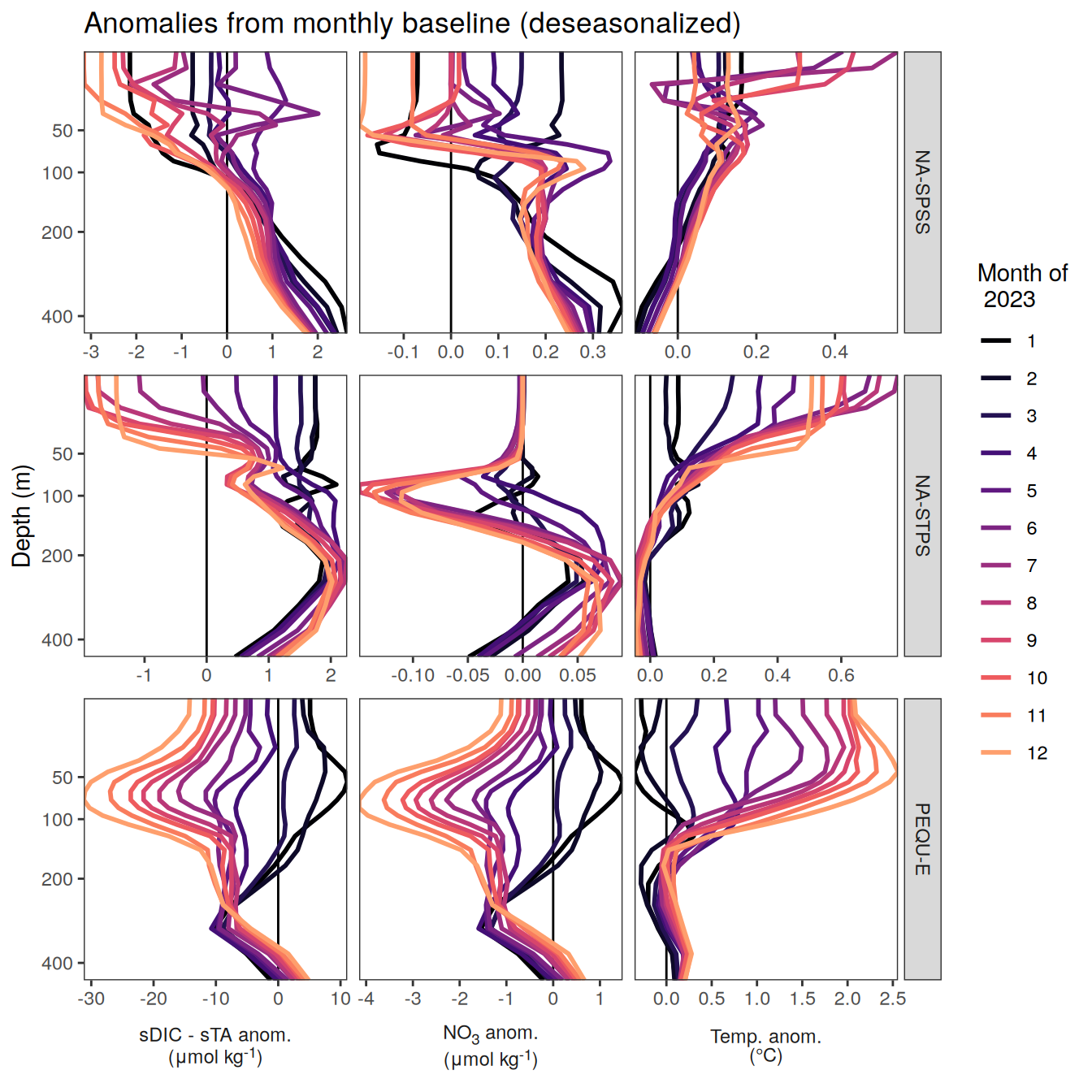
| Version | Author | Date |
|---|---|---|
| 945d8f7 | jens-daniel-mueller | 2025-02-23 |
| c50054d | jens-daniel-mueller | 2024-08-29 |
| ba4aaac | jens-daniel-mueller | 2024-07-08 |
| 197dac4 | jens-daniel-mueller | 2024-06-27 |
| f03b1d8 | jens-daniel-mueller | 2024-06-12 |
| de65385 | jens-daniel-mueller | 2024-06-12 |
| 34b4fe2 | jens-daniel-mueller | 2024-06-12 |
| 0a7394b | jens-daniel-mueller | 2024-06-11 |
| 54af933 | jens-daniel-mueller | 2024-06-03 |
# ggsave(width = 10,
# height = 8,
# dpi = 600,
# filename = "../output/FESOM_2023_anomaly_profiles.jpg")Hovmoeller
plot_list <-
full_join(
pco2_product_profiles_monthly %>%
filter(
year == 2023,
biome %in% key_biomes,
name %in% c("sdissic_stalk", "thetao")
),
pco2_product_biome_monthly_detrended %>%
filter(
biome %in% key_biomes,
name %in% "mld",
year == 2023,
product %in% gobm_product_list
) %>%
select(product, month, biome, mld = value)
) %>%
group_split(name, biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_contour_filled(aes(month, depth, z = resid)) +
geom_line(aes(month, mld))+
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
super = ScaleDiscretised,
name = labels_breaks(.x %>% distinct(name))$i_legend_title
)+
scale_y_continuous(trans = trans_reverser("sqrt"), breaks = c(20, 50, 100, 200, 400)) +
coord_cartesian(expand = 0,
ylim = c(300,NA)) +
facet_grid(product ~ biome) +
guides(
fill = guide_colorsteps(
barheight = unit(0.3, "cm"),
barwidth = unit(10, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.position = "top",
legend.title.align = 1,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(halign = 1,
lineheight = 1.5)
)
)
plot_list[[1]]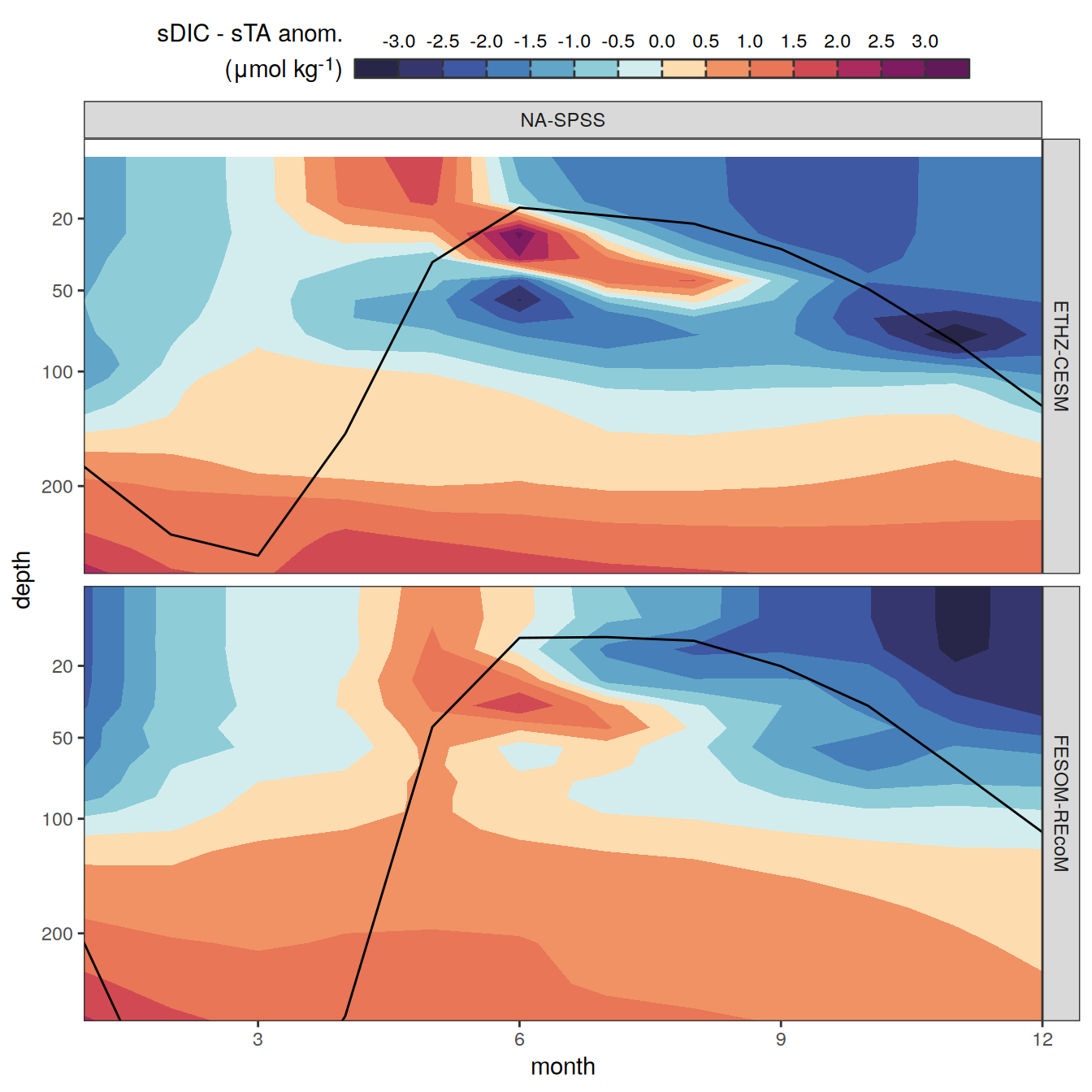
[[2]]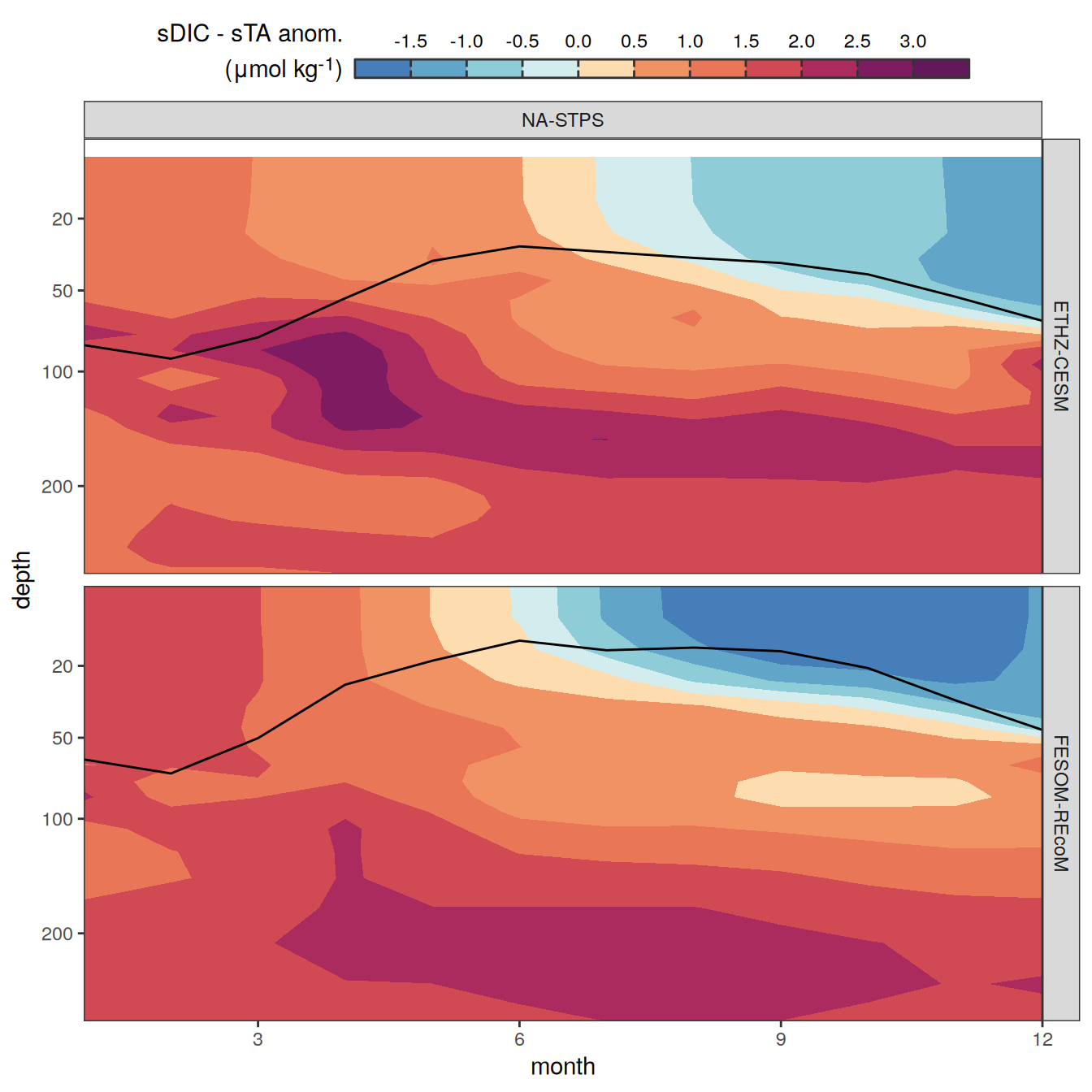
[[3]]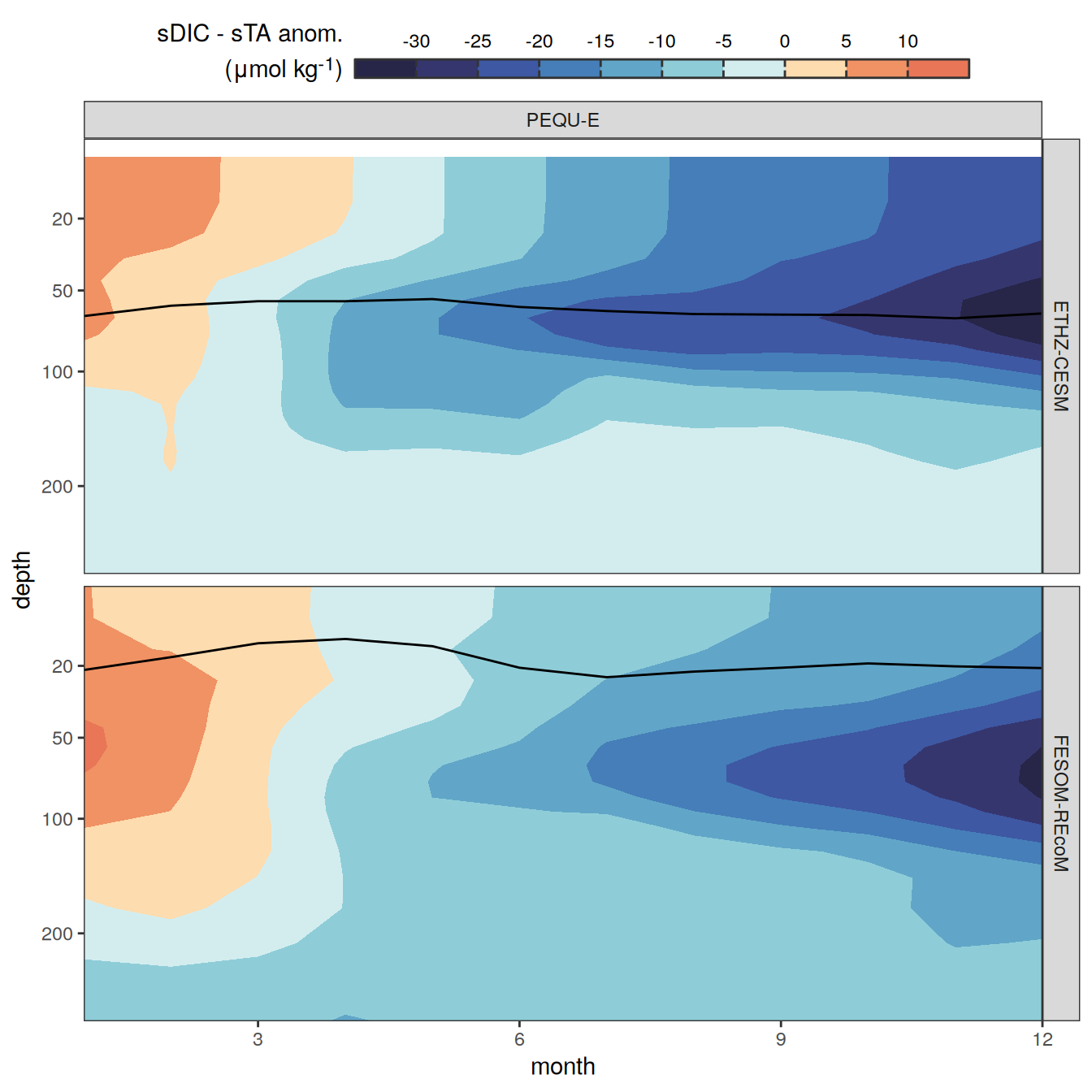
[[4]]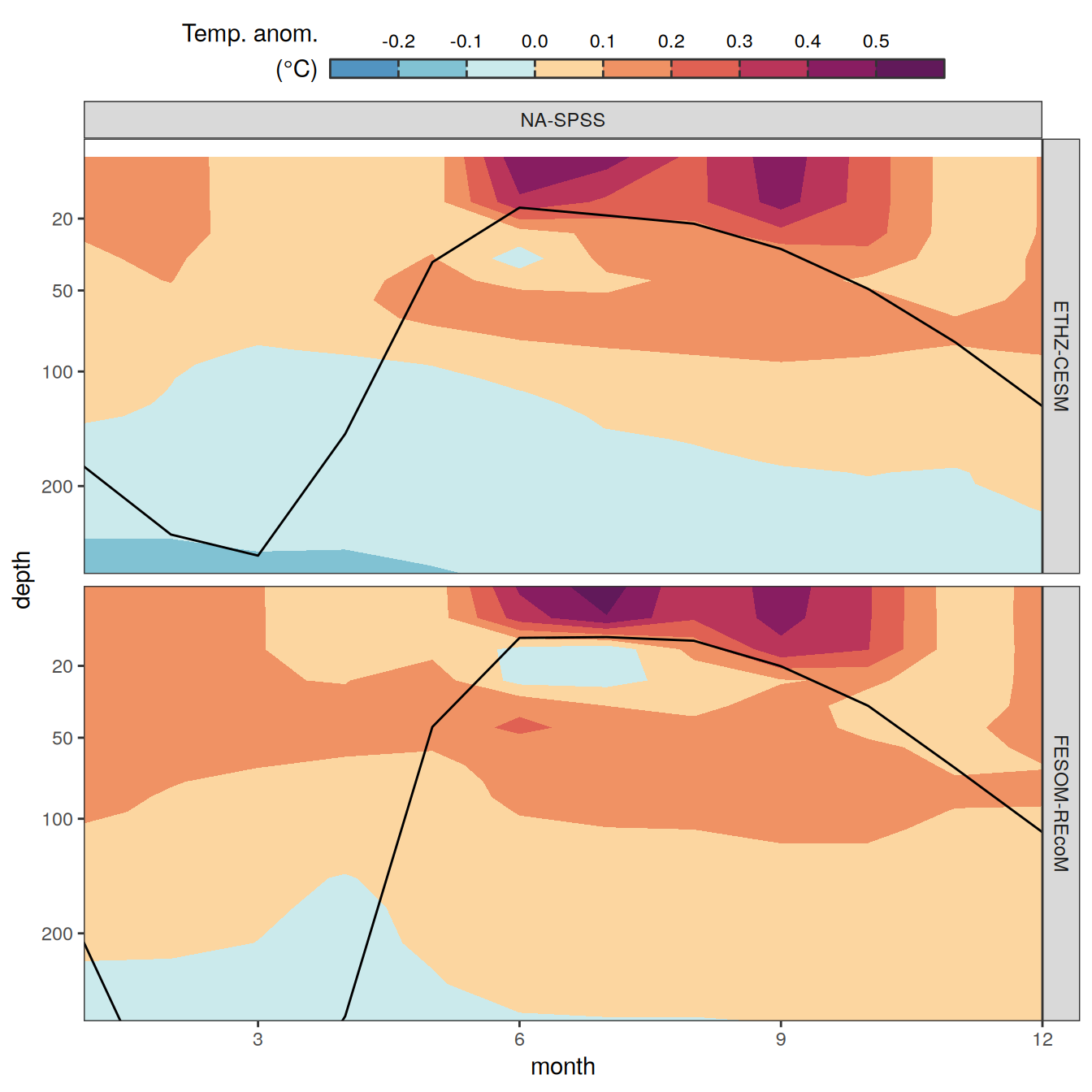
[[5]]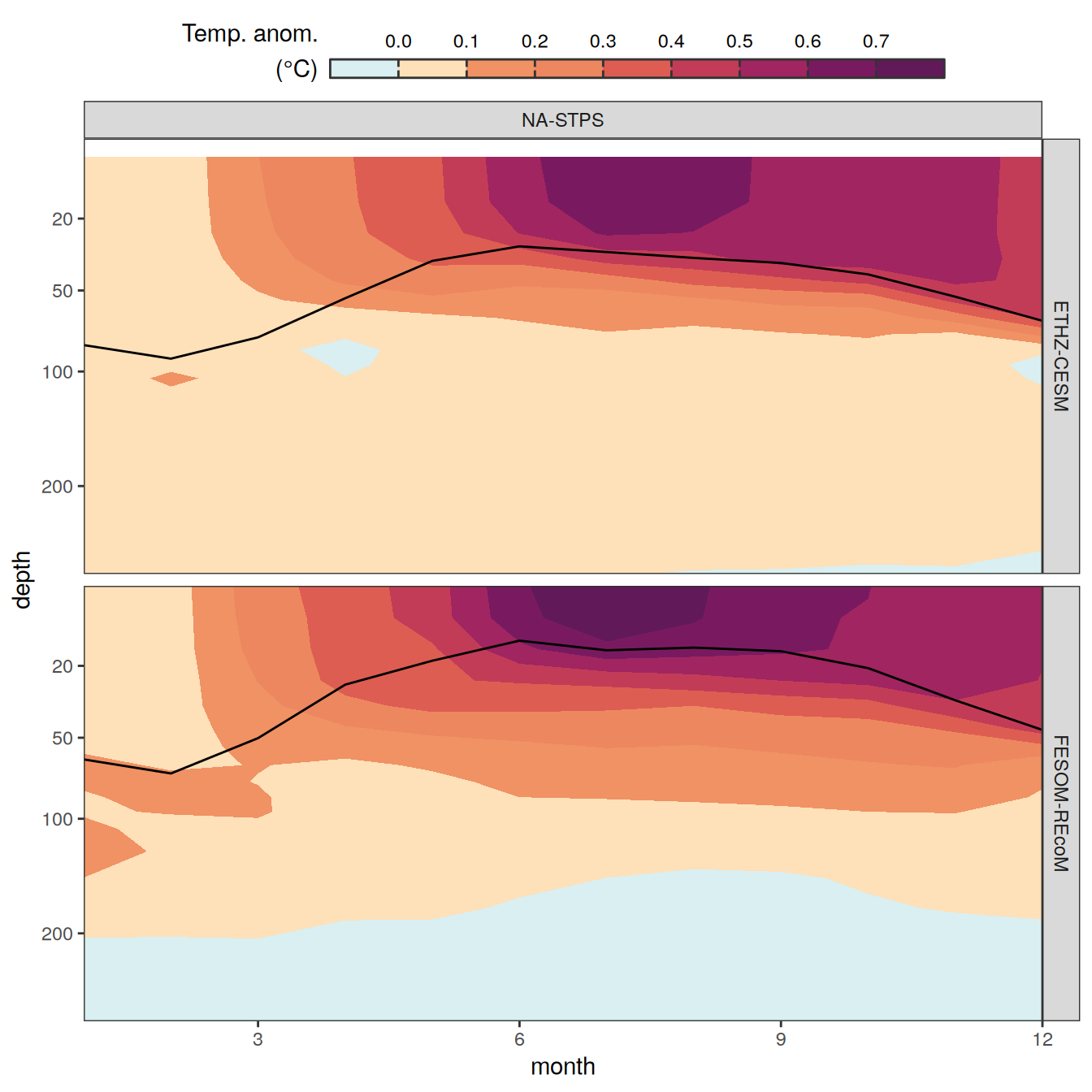
[[6]]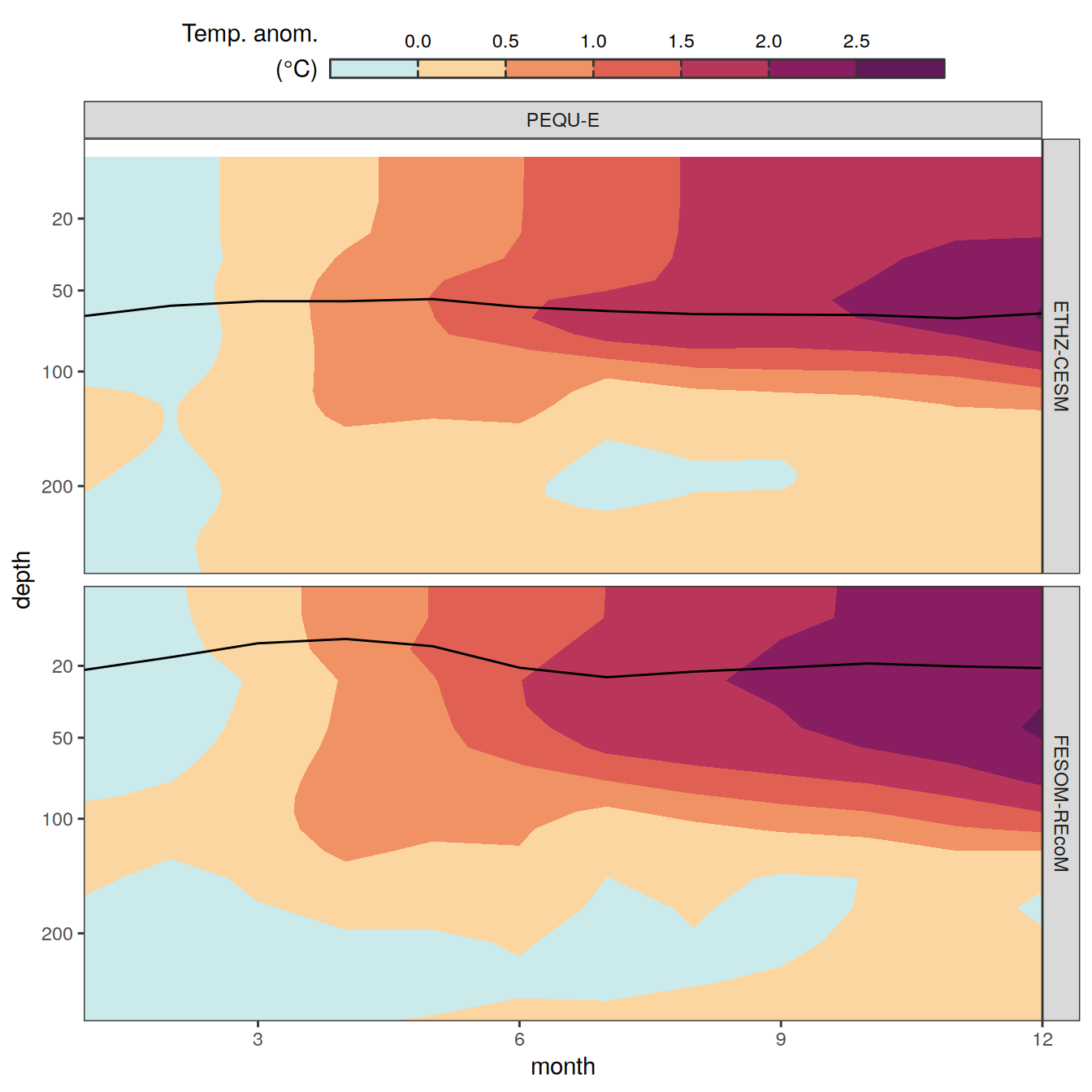
ggsave(plot = wrap_plots(plot_list,
ncol = 3),
width = 18,
height = 12,
dpi = 600,
filename = "../output/profiles_hovmoeller_all_gobm.jpg")
plot_list <-
full_join(
pco2_product_profiles_monthly %>%
filter(
year == 2023,
biome %in% key_biomes,
name %in% c("sdissic_stalk", "thetao")
) %>%
arrange(month) %>%
group_by(product, name, biome, depth) %>%
mutate(resid = if_else(name == "sdissic_stalk",
resid - first(resid),
resid)) %>%
ungroup(),
pco2_product_biome_monthly_detrended %>%
filter(
biome %in% key_biomes,
name %in% "mld",
year == 2023,
product %in% gobm_product_list
) %>%
select(product, month, biome, mld = value)
) %>%
group_split(name, biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_contour_filled(aes(month, depth, z = resid)) +
geom_line(aes(month, mld))+
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
super = ScaleDiscretised,
name = labels_breaks(.x %>% distinct(name))$i_legend_title
)+
scale_y_continuous(trans = trans_reverser("sqrt"), breaks = c(20, 50, 100, 200, 400)) +
coord_cartesian(expand = 0,
ylim = c(300,NA)) +
facet_grid(product ~ biome) +
guides(
fill = guide_colorsteps(
barheight = unit(0.3, "cm"),
barwidth = unit(10, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.position = "top",
legend.title.align = 1,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(halign = 1,
lineheight = 1.5)
)
)
ggsave(plot = wrap_plots(plot_list,
ncol = 3),
width = 18,
height = 12,
dpi = 600,
filename = "../output/profiles_hovmoeller_all_gobm_evolution.jpg")CESM_depth_grid <- pco2_product_profiles_monthly %>%
filter(year == 2023,
product == "ETHZ-CESM",
biome %in% key_biomes,
name %in% c("sdissic_stalk", "thetao")) %>%
distinct(name, biome, month, depth)
pco2_product_profiles_monthly_FESOM_regrid <-
full_join(
pco2_product_profiles_monthly %>%
filter(
year == 2023,
product == "FESOM-REcoM",
biome %in% key_biomes,
name %in% c("sdissic_stalk", "thetao")
),
CESM_depth_grid %>% mutate(product = "FESOM-REcoM")
)
pco2_product_profiles_monthly_FESOM_regrid <-
pco2_product_profiles_monthly_FESOM_regrid %>%
arrange(product, name, biome, month, depth)
pco2_product_profiles_monthly_FESOM_regrid <-
pco2_product_profiles_monthly_FESOM_regrid %>%
arrange(depth) %>%
group_by(product, name, biome, month) %>%
mutate(resid = spline(
depth,
resid,
method = "natural",
xout = depth
)$y) %>%
ungroup()
CESM_depth <-
CESM_depth_grid %>% distinct(depth) %>% pull()
pco2_product_profiles_monthly_FESOM_regrid <-
pco2_product_profiles_monthly_FESOM_regrid %>%
filter(depth %in% CESM_depth)
pco2_product_profiles_monthly_merged <-
bind_rows(
pco2_product_profiles_monthly_FESOM_regrid,
pco2_product_profiles_monthly %>%
filter(
year == 2023,
product == "ETHZ-CESM",
biome %in% key_biomes,
name %in% c("sdissic_stalk", "thetao")
)
)
pco2_product_profiles_monthly_ensemble <-
pco2_product_profiles_monthly_merged %>%
group_by(name, biome, month, depth) %>%
summarise(resid = mean(resid)) %>%
ungroup()
pco2_product_profiles_monthly_ensemble <-
full_join(
pco2_product_profiles_monthly_ensemble %>%
filter(
biome %in% key_biomes,
name %in% c("sdissic_stalk", "thetao")
),
pco2_product_biome_monthly_detrended %>%
filter(
biome %in% key_biomes,
name %in% "mld",
year == 2023,
product %in% gobm_product_list
) %>%
group_by(month, biome) %>%
summarise(mld = mean(value)) %>%
ungroup()
)
# plot_list <-
pco2_product_profiles_monthly_ensemble %>%
group_split(name, biome) %>%
head(1) %>%
map(
~ ggplot(data = .x) +
geom_contour_filled(aes(month, depth, z = resid)) +
geom_line(aes(month, mld)) +
scale_fill_gradientn(
colours = warm_cool_gradient,
rescaler = ~ scales::rescale_mid(.x, mid = 0),
super = ScaleDiscretised,
name = labels_breaks(.x %>% distinct(name))$i_legend_title
) +
scale_y_continuous(trans = trans_reverser("sqrt"), breaks = c(50, 100, 200, 400)) +
coord_cartesian(expand = 0, ylim = c(300, NA)) +
facet_wrap(~ biome) +
guides(
fill = guide_colorsteps(
barheight = unit(0.3, "cm"),
barwidth = unit(10, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.position = "top",
legend.title.align = 1,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(halign = 1, lineheight = 1.5)
)
)[[1]]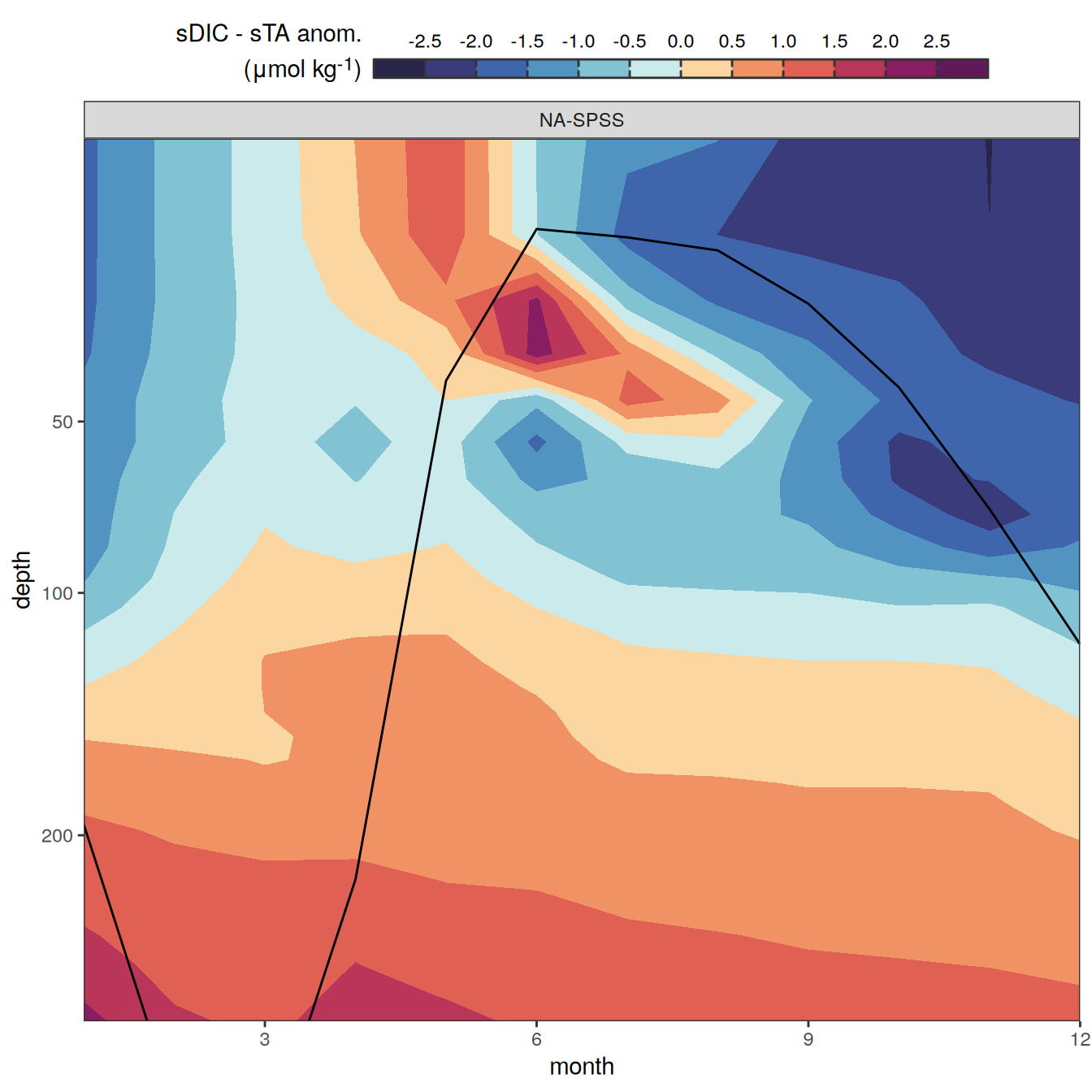
ggsave(plot = wrap_plots(plot_list,
ncol = 3),
width = 18,
height = 8,
dpi = 600,
filename = "../output/profiles_hovmoeller_ensemble_mean_gobm.jpg")labels_breaks_hov <- function(i_name, i_biome) {
if (i_name == "sdissic_stalk") {
i_legend_title <- "sDIC - sTA<br>anom.<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "thetao") {
i_legend_title <- "Temp.<br>anom.<br>(°C)"
}
if (i_name == "sdissic_stalk" & i_biome == "NA-SPSS") {
i_breaks <- c(-Inf, seq(-2, 2, 0.5), Inf)
}
if (i_name == "thetao" & i_biome == "NA-SPSS") {
i_breaks <- c(-Inf, seq(-0.4, 0.4, 0.1), Inf)
}
if (i_name == "sdissic_stalk" & i_biome == "NA-STPS") {
i_breaks <- c(-Inf, seq(-2.4, 2.4, 0.6), Inf)
}
if (i_name == "thetao" & i_biome == "NA-STPS") {
i_breaks <- c(-Inf, seq(-0.6, 0.6, 0.15), Inf)
}
if (i_name == "sdissic_stalk" & i_biome == "PEQU-E") {
i_breaks <- c(-Inf, seq(-32, 32, 8), Inf)
}
if (i_name == "thetao" & i_biome == "PEQU-E") {
i_breaks <- c(-Inf, seq(-2, 2, 0.5), Inf)
}
i_breaks_labels <- i_breaks[!i_breaks == Inf]
i_breaks_labels <- i_breaks_labels[!i_breaks_labels == -Inf]
i_breaks_labels[seq_along(i_breaks_labels) %% 2 == 0] <- ""
all_labels_breaks <- lst(i_legend_title, i_breaks, i_breaks_labels)
return(all_labels_breaks)
}
labels_breaks_hov("sdissic_stalk", "NA-SPSS")$i_legend_title
[1] "sDIC - sTA<br>anom.<br>(μmol kg<sup>-1</sup>)"
$i_breaks
[1] -Inf -2.0 -1.5 -1.0 -0.5 0.0 0.5 1.0 1.5 2.0 Inf
$i_breaks_labels
[1] "-2" "" "-1" "" "0" "" "1" "" "2" plot_list_left <-
pco2_product_profiles_monthly_ensemble %>%
arrange(month) %>%
group_by(name, biome, depth) %>%
mutate(resid = if_else(name == "sdissic_stalk", resid - first(resid), resid)) %>%
ungroup() %>%
group_split(biome, name) %>%
head(2) %>%
map(
~ ggplot(data = .x) +
geom_contour_filled(aes(month, depth, z = resid),
breaks = labels_breaks_hov(.x %>% distinct(name),
.x %>% distinct(biome))$i_breaks) +
geom_line(aes(month, mld)) +
scale_fill_gradientn(
colours = warm_cool_gradient,
super = ScaleDiscretised,
name = labels_breaks_hov(.x %>% distinct(name),
.x %>% distinct(biome))$i_legend_title,
labels = labels_breaks_hov(.x %>% distinct(name),
.x %>% distinct(biome))$i_breaks_labels
) +
# scale_fill_gradientn(
# colours = warm_cool_gradient,
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# super = ScaleDiscretised,
# name = labels_breaks(.x %>% distinct(name))$i_legend_title
# ) +
scale_y_continuous(
trans = trans_reverser("sqrt"),
breaks = c(20, 50, 100, 200, 400)
) +
coord_cartesian(expand = 0, ylim = c(300, NA)) +
labs(y = "Depth (m)",
x = "Month") +
facet_wrap(~ biome) +
guides(
fill = guide_colorsteps(
barheight = unit(0.3, "cm"),
barwidth = unit(5, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.position = "top",
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_markdown(hjust = 1,
lineheight = 1.5)
)
)
plot_list_right <-
pco2_product_profiles_monthly_ensemble %>%
arrange(month) %>%
group_by(name, biome, depth) %>%
mutate(resid = if_else(name == "sdissic_stalk", resid - first(resid), resid)) %>%
ungroup() %>%
group_split(biome, name) %>%
tail(4) %>%
map(
~ ggplot(data = .x) +
geom_contour_filled(aes(month, depth, z = resid),
breaks = labels_breaks_hov(.x %>% distinct(name),
.x %>% distinct(biome))$i_breaks) +
geom_line(aes(month, mld)) +
scale_fill_gradientn(
colours = warm_cool_gradient,
super = ScaleDiscretised,
name = labels_breaks_hov(.x %>% distinct(name),
.x %>% distinct(biome))$i_legend_title,
labels = labels_breaks_hov(.x %>% distinct(name),
.x %>% distinct(biome))$i_breaks_labels
) +
scale_y_continuous(
trans = trans_reverser("sqrt"),
breaks = c(20, 50, 100, 200, 400)
) +
coord_cartesian(expand = 0, ylim = c(300, NA)) +
labs(y = "Depth (m)", x = "Month")+
facet_wrap(~ biome) +
guides(
fill = guide_colorsteps(
barheight = unit(0.3, "cm"),
barwidth = unit(5, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.position = "top",
# legend.margin = margin(0, 0, 0, 0),
# legend.justification = "left",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
legend.title.align = 1,
legend.box.spacing = unit(0.1, "cm"),
legend.title = element_blank()
)
)
plot_list <- c(plot_list_left, plot_list_right)
ggsave(plot = wrap_plots(plot_list,
ncol = 3,
byrow = FALSE),
width = 10,
height = 6,
dpi = 600,
filename = "../output/profiles_hovmoeller_ensemble_mean_gobm_evolution.jpg")
sessionInfo()R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: openSUSE Leap 15.6
Matrix products: default
BLAS/LAPACK: /usr/local/OpenBLAS-0.3.28/lib/libopenblas_haswellp-r0.3.28.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Zurich
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] seacarb_3.3.3 SolveSAPHE_2.1.0 oce_1.8-3
[4] gsw_1.2-0 kableExtra_1.4.0 cmocean_0.3-2
[7] ggh4x_0.3.0 scales_1.3.0 biscale_1.0.0
[10] ggtext_0.1.2 khroma_1.14.0 ggnewscale_0.5.0
[13] terra_1.8-5 sf_1.0-19 rnaturalearth_1.0.1
[16] geomtextpath_0.1.4 colorspace_2.1-1 marelac_2.1.11
[19] shape_1.4.6.1 ggforce_0.4.2 metR_0.16.0
[22] scico_1.5.0 patchwork_1.3.0 collapse_2.0.18
[25] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
[28] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5
[31] tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.1
[34] tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] DBI_1.2.3 rlang_1.1.4 magrittr_2.0.3
[4] git2r_0.35.0 e1071_1.7-16 compiler_4.4.2
[7] mgcv_1.9-1 getPass_0.2-4 systemfonts_1.1.0
[10] callr_3.7.6 vctrs_0.6.5 pkgconfig_2.0.3
[13] crayon_1.5.3 fastmap_1.2.0 backports_1.5.0
[16] labeling_0.4.3 utf8_1.2.4 promises_1.3.2
[19] rmarkdown_2.29 markdown_1.13 tzdb_0.4.0
[22] ps_1.8.1 ragg_1.3.3 bit_4.5.0
[25] xfun_0.49 cachem_1.1.0 jsonlite_1.8.9
[28] later_1.4.1 tweenr_2.0.3 parallel_4.4.2
[31] R6_2.5.1 RColorBrewer_1.1-3 bslib_0.8.0
[34] stringi_1.8.4 jquerylib_0.1.4 Rcpp_1.0.13-1
[37] knitr_1.49 Matrix_1.7-1 splines_4.4.2
[40] httpuv_1.6.15 timechange_0.3.0 tidyselect_1.2.1
[43] rstudioapi_0.17.1 yaml_2.3.10 codetools_0.2-20
[46] processx_3.8.4 lattice_0.22-6 withr_3.0.2
[49] evaluate_1.0.1 isoband_0.2.7 rnaturalearthdata_1.0.0
[52] units_0.8-5 proxy_0.4-27 polyclip_1.10-7
[55] xml2_1.3.6 pillar_1.9.0 whisker_0.4.1
[58] KernSmooth_2.23-24 checkmate_2.3.2 generics_0.1.3
[61] vroom_1.6.5 rprojroot_2.0.4 hms_1.1.3
[64] commonmark_1.9.2 munsell_0.5.1 class_7.3-22
[67] glue_1.8.0 tools_4.4.2 data.table_1.16.2
[70] fs_1.6.5 cowplot_1.1.3 grid_4.4.2
[73] nlme_3.1-166 cli_3.6.3 textshaping_0.4.0
[76] fansi_1.0.6 viridisLite_0.4.2 svglite_2.1.3
[79] gtable_0.3.6 sass_0.4.9 digest_0.6.37
[82] classInt_0.4-10 farver_2.1.2 memoise_2.0.1
[85] htmltools_0.5.8.1 lifecycle_1.0.4 httr_1.4.7
[88] here_1.0.1 gridtext_0.1.5 bit64_4.5.2
[91] MASS_7.3-61