NRT_fco2residual
Jens Daniel Müller
07 March, 2025
Last updated: 2025-03-07
Checks: 7 0
Knit directory:
heatwave_co2_flux_2023/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240307) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version dd25c0c. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data
Ignored: output/
Unstaged changes:
Modified: analysis/_site.yml
Modified: analysis/child/pCO2_product_synopsis.Rmd
Modified: code/Workflowr_project_managment.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/NRT_fco2residual.Rmd) and
HTML (docs/NRT_fco2residual.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 2b1d73c | jens-daniel-mueller | 2025-03-01 | Build site. |
| html | dc38c97 | jens-daniel-mueller | 2025-02-26 | Build site. |
| html | 2ea5f0a | jens-daniel-mueller | 2025-02-25 | Build site. |
| html | 698803c | jens-daniel-mueller | 2025-02-25 | Build site. |
| html | f7331c3 | jens-daniel-mueller | 2025-02-23 | Build site. |
| html | 06d9c78 | jens-daniel-mueller | 2025-02-23 | Build site. |
| html | 1ca18ff | jens-daniel-mueller | 2025-02-22 | Build site. |
| html | 2636a99 | jens-daniel-mueller | 2025-02-21 | Build site. |
| html | 551f825 | jens-daniel-mueller | 2024-09-09 | Build site. |
| html | 7c52bc2 | jens-daniel-mueller | 2024-08-23 | Build site. |
| html | 4f019e4 | jens-daniel-mueller | 2024-07-11 | Build site. |
| html | 334ff26 | jens-daniel-mueller | 2024-07-10 | manual commit |
| html | 430e926 | jens-daniel-mueller | 2024-07-10 | manual commit |
| html | 3c01ce1 | jens-daniel-mueller | 2024-07-08 | Build site. |
| Rmd | 59699d6 | jens-daniel-mueller | 2024-07-08 | rename fCO2-Residual |
| html | f6a4369 | jens-daniel-mueller | 2024-07-01 | Build site. |
| html | f1954bc | jens-daniel-mueller | 2024-06-27 | Build site. |
| html | a039cda | jens-daniel-mueller | 2024-06-26 | Build site. |
| html | 1a3d3c8 | jens-daniel-mueller | 2024-06-13 | Build site. |
| html | 3148fef | jens-daniel-mueller | 2024-06-13 | Build site. |
| html | 3969e52 | jens-daniel-mueller | 2024-06-12 | Build site. |
| html | a60be97 | jens-daniel-mueller | 2024-06-12 | Build site. |
| Rmd | 02feef8 | jens-daniel-mueller | 2024-06-12 | free memory |
| html | d46002d | jens-daniel-mueller | 2024-06-12 | manual commit |
| html | c69cb32 | jens-daniel-mueller | 2024-06-11 | Build site. |
| html | 5261667 | jens-daniel-mueller | 2024-06-11 | manual commit |
| html | 6954c65 | jens-daniel-mueller | 2024-06-06 | Build site. |
| html | e1e0ccb | jens-daniel-mueller | 2024-05-27 | Build site. |
| Rmd | d8f416a | jens-daniel-mueller | 2024-05-27 | consistency fixes |
| html | a3743ec | jens-daniel-mueller | 2024-05-25 | Build site. |
| Rmd | f1253fd | jens-daniel-mueller | 2024-05-25 | SO processed, but not included in global integrals |
| html | be285dc | jens-daniel-mueller | 2024-05-21 | Build site. |
| html | 51df30d | jens-daniel-mueller | 2024-05-15 | Build site. |
| Rmd | 981d5e1 | jens-daniel-mueller | 2024-05-15 | kw K0 product included, mean flux densities computed |
| html | 009791f | jens-daniel-mueller | 2024-05-14 | Build site. |
| html | 3b5d16b | jens-daniel-mueller | 2024-05-13 | Build site. |
| Rmd | 1e1dee5 | jens-daniel-mueller | 2024-05-13 | pco2 to fco2 conversions, changed output files |
| html | 1ea301d | jens-daniel-mueller | 2024-05-06 | Build site. |
| Rmd | d26f47c | jens-daniel-mueller | 2024-05-06 | use new version of OceanSODA, unit error fixed in Residual |
| html | cd94a01 | jens-daniel-mueller | 2024-05-06 | Build site. |
| Rmd | 2ea0165 | jens-daniel-mueller | 2024-05-06 | use version 5may2024 |
| html | 7f9c687 | jens-daniel-mueller | 2024-04-23 | Build site. |
| Rmd | 246f9cd | jens-daniel-mueller | 2024-04-19 | manual commit |
| html | 231f7cd | jens-daniel-mueller | 2024-04-17 | Build site. |
| Rmd | 20624c0 | jens-daniel-mueller | 2024-04-17 | rebuild with corrected NRT units |
| html | ce4e2a6 | jens-daniel-mueller | 2024-04-17 | Build site. |
| Rmd | 6b3a080 | jens-daniel-mueller | 2024-04-17 | rebuild entire website with NRT_fco2residual |
| html | 741ee62 | jens-daniel-mueller | 2024-04-17 | Build site. |
| Rmd | b7cc891 | jens-daniel-mueller | 2024-04-17 | ingest new product, rerun SOM-FFN |
| html | 3bd3bfb | jens-daniel-mueller | 2024-04-17 | Build site. |
| Rmd | 31deaad | jens-daniel-mueller | 2024-04-17 | ingest new product |
center <- -160
boundary <- center + 180
target_crs <- paste0("+proj=robin +over +lon_0=", center)
# target_crs <- paste0("+proj=eqearth +over +lon_0=", center)
# target_crs <- paste0("+proj=eqearth +lon_0=", center)
# target_crs <- paste0("+proj=igh_o +lon_0=", center)
worldmap <- ne_countries(scale = 'small',
type = 'map_units',
returnclass = 'sf')
worldmap <- worldmap %>% st_break_antimeridian(lon_0 = center)
worldmap_trans <- st_transform(worldmap, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans)
coastline <- ne_coastline(scale = 'small', returnclass = "sf")
coastline <- st_break_antimeridian(coastline, lon_0 = 200)
coastline_trans <- st_transform(coastline, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col="grey") +
# geom_sf(data = coastline_trans)
bbox <- st_bbox(c(xmin = -180, xmax = 180, ymax = 65, ymin = -78), crs = st_crs(4326))
bbox <- st_as_sfc(bbox)
bbox_trans <- st_break_antimeridian(bbox, lon_0 = center)
bbox_graticules <- st_graticule(
x = bbox_trans,
crs = st_crs(bbox_trans),
datum = st_crs(bbox_trans),
lon = c(20, 20.001),
lat = c(-78,65),
ndiscr = 1e3,
margin = 0.001
)
bbox_graticules_trans <- st_transform(bbox_graticules, crs = target_crs)
rm(worldmap, coastline, bbox, bbox_trans)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col="grey") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans)
lat_lim <- ext(bbox_graticules_trans)[c(3,4)]*1.002
lon_lim <- ext(bbox_graticules_trans)[c(1,2)]*1.005
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey90", col = "grey90") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans, linewidth = 1) +
# coord_sf(crs = target_crs,
# ylim = lat_lim,
# xlim = lon_lim,
# expand = FALSE) +
# theme(
# panel.border = element_blank(),
# axis.text = element_blank(),
# axis.ticks = element_blank()
# )
latitude_graticules <- st_graticule(
x = bbox_graticules,
crs = st_crs(bbox_graticules),
datum = st_crs(bbox_graticules),
lon = c(20, 20.001),
lat = c(-60,-30,0,30,60),
ndiscr = 1e3,
margin = 0.001
)
latitude_graticules_trans <- st_transform(latitude_graticules, crs = target_crs)
latitude_labels <- data.frame(lat_label = c("60°N","30°N","Eq.","30°S","60°S"),
lat = c(60,30,0,-30,-60)-4, lon = c(35)-c(0,2,4,2,0))
latitude_labels <- st_as_sf(x = latitude_labels,
coords = c("lon", "lat"),
crs = "+proj=longlat")
latitude_labels_trans <- st_transform(latitude_labels, crs = target_crs)
# ggplot() +
# geom_sf(data = worldmap_trans, fill = "grey", col = "grey") +
# geom_sf(data = coastline_trans) +
# geom_sf(data = bbox_graticules_trans) +
# geom_sf(data = latitude_graticules_trans,
# col = "grey60",
# linewidth = 0.2) +
# geom_sf_text(data = latitude_labels_trans,
# aes(label = lat_label),
# size = 3,
# col = "grey60")Read data
path_pCO2_products <-
"/nfs/kryo/work/datasets/gridded/ocean/2d/observation/pco2/"
path_NRT_fco2residual <-
"/nfs/kryo/work/datasets/gridded/ocean/2d/observation/pco2/NRT_fco2residual_mckinley/"library(ncdf4)
nc <-
nc_open(paste0(
path_pCO2_products,
"VLIZ-SOM_FFN/VLIZ-SOM_FFN_vBAMS2024.nc"
))
nc <-
nc_open(paste0(
path_pCO2_products,
"VLIZ-SOM_FFN/VLIZ-SOM_FFN_inputs.nc"
))
nc <-
nc_open(paste0(
path_NRT_fco2residual,
"NRT_fco2residual_mckinley_5may2024.nc"
))
print(nc)print("NRT_fco2residual_mckinley_5may2024.nc")[1] "NRT_fco2residual_mckinley_5may2024.nc"pco2_product <-
read_ncdf(
paste0(
path_NRT_fco2residual,
"NRT_fco2residual_mckinley_5may2024.nc"
),
make_units = FALSE
)
pco2_product <- pco2_product %>%
as_tibble()
pco2_product <- pco2_product %>%
select(-mld)
pco2_product <-
pco2_product %>%
rename(lon = xlon,
lat = ylat,
atm_fco2 = fco2atm,
sol = alpha,
salinity = sos,
temperature = tos)
pco2_product <-
pco2_product %>%
mutate(#fgco2 = fgco2 * 60 * 60 * 24 * 365 * 1e-14,
dfco2 = sfco2 - atm_fco2,
kw = kw * 1e-2 * 24 * 365,
sol = sol * 1e-3,
chl = log10(chl))
pco2_product <-
pco2_product %>%
mutate(kw_sol = kw * sol) %>%
select(-c(kw, sol))
# pco2_product %>%
# ggplot(aes(sol)) +
# geom_histogram()
# pco2_product %>%
# filter(mld < 50) %>%
# ggplot(aes(mld)) +
# geom_histogram()
# pco2_product <-
# pco2_product %>%
# mutate(across(mld, ~ replace(., . == 0, NA)))
pco2_product <-
pco2_product %>%
mutate(area = earth_surf(lat, lon),
year = year(time),
month = month(time))
# pco2_product %>%
# filter(year == 2023,
# month %in% c(11,12)) %>%
# ggplot(aes(lon, lat, fill = fgco2)) +
# geom_tile() +
# facet_wrap(~ month)
pco2_product <-
pco2_product %>%
mutate(lon = if_else(lon < 20, lon + 360, lon))pCO2_product_preprocessing <-
knitr::knit_expand(
file = here::here("analysis/child/pCO2_product_preprocessing.Rmd"),
product_name = "fCO2-Residual"
)Preprocessing
# model <- TRUE
model <- str_detect('fCO2-Residual', "FESOM-REcoM|ETHZ-CESM")Load masks
biome_mask <-
read_rds(here::here("data/biome_mask.rds"))
region_mask <-
read_rds(here::here("data/region_mask.rds"))
map <-
read_rds(here::here("data/map.rds"))
key_biomes <-
read_rds(here::here("data/key_biomes.rds"))Define labels and breaks
labels_breaks <- function(i_name) {
if (i_name == "dco2") {
i_legend_title <- "ΔpCO<sub>2</sub><br>(µatm)"
}
if (i_name == "dfco2") {
i_legend_title <- "ΔfCO<sub>2</sub><br>(µatm)"
}
if (i_name == "atm_co2") {
i_legend_title <- "pCO<sub>2,atm</sub><br>(µatm)"
}
if (i_name == "atm_fco2") {
i_legend_title <- "fCO<sub>2,atm</sub><br>(µatm)"
}
if (i_name == "sol") {
i_legend_title <- "K<sub>0</sub><br>(mol m<sup>-3</sup> µatm<sup>-1</sup>)"
}
if (i_name == "kw") {
i_legend_title <- "k<sub>w</sub><br>(m yr<sup>-1</sup>)"
}
if (i_name == "kw_sol") {
i_legend_title <- "k<sub>w</sub> K<sub>0</sub><br>(mol yr<sup>-1</sup> m<sup>-2</sup> µatm<sup>-1</sup>)"
}
if (i_name == "spco2") {
i_legend_title <- "pCO<sub>2,ocean</sub><br>(µatm)"
}
if (i_name == "sfco2") {
i_legend_title <- "fCO<sub>2,ocean</sub><br>(µatm)"
}
if (i_name == "intpp") {
i_legend_title <- "NPP<sub>int</sub><br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
}
if (i_name == "no3") {
i_legend_title <- "NO<sub>3</sub><br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "o2") {
i_legend_title <- "O<sub>2</sub><br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "dissic") {
i_legend_title <- "DIC<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "sdissic") {
i_legend_title <- "sDIC<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "cstar") {
i_legend_title <- "C*<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "talk") {
i_legend_title <- "TA<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "stalk") {
i_legend_title <- "sTA<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "sdissic_stalk") {
i_legend_title <- "sDIC-sTA<br>(μmol kg<sup>-1</sup>)"
}
if (i_name == "sfco2_total") {
i_legend_title <- "total"
}
if (i_name == "sfco2_therm") {
i_legend_title <- "thermal"
}
if (i_name == "sfco2_nontherm") {
i_legend_title <- "non-thermal"
}
if (i_name == "fgco2") {
i_legend_title <- "FCO<sub>2</sub><br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
}
if (i_name == "fgco2_predict") {
i_legend_title <- "FCO<sub>2</sub> pred.<br>(mol m<sup>-2</sup> yr<sup>-1</sup>)"
}
if (i_name == "fgco2_hov") {
i_legend_title <- "FCO<sub>2</sub><br>(PgC deg<sup>-1</sup> yr<sup>-1</sup>)"
}
if (i_name == "fgco2_int") {
i_legend_title <- "FCO<sub>2</sub><br>(PgC yr<sup>-1</sup>)"
}
if (i_name == "fgco2_predict_int") {
i_legend_title <- "FCO<sub>2</sub> pred.<br>(PgC yr<sup>-1</sup>)"
}
if (i_name == "thetao") {
i_legend_title <- "Temp.<br>(°C)"
}
if (i_name == "temperature") {
i_legend_title <- "SST<br>(°C)"
}
if (i_name == "salinity") {
i_legend_title <- "SSS"
}
if (i_name == "so") {
i_legend_title <- "salinity"
}
if (i_name == "chl") {
i_legend_title <- "lg(Chl-a)<br>(lg(mg m<sup>-3</sup>))"
}
if (i_name == "mld") {
i_legend_title <- "MLD<br>(m)"
}
if (i_name == "press") {
i_legend_title <- "pressure<sub>atm</sub><br>(Pa)"
}
if (i_name == "wind") {
i_legend_title <- "Wind <br>(m sec<sup>-1</sup>)"
}
if (i_name == "SSH") {
i_legend_title <- "SSH <br>(m)"
}
if (i_name == "fice") {
i_legend_title <- "Sea ice <br>(%)"
}
if (i_name == "resid_fgco2") {
i_legend_title <-
"Observed"
}
if (i_name == "resid_fgco2_dfco2") {
i_legend_title <-
"ΔfCO<sub>2</sub>"
}
if (i_name == "resid_fgco2_kw_sol") {
i_legend_title <-
"k<sub>w</sub> K<sub>0</sub>"
}
if (i_name == "resid_fgco2_dfco2_kw_sol") {
i_legend_title <-
"k<sub>w</sub> K<sub>0</sub> X ΔfCO<sub>2</sub>"
}
if (i_name == "resid_fgco2_sum") {
i_legend_title <-
"∑"
}
if (i_name == "resid_fgco2_offset") {
i_legend_title <-
"Obs. - ∑"
}
all_labels_breaks <- lst(i_legend_title)
return(all_labels_breaks)
}
x_axis_labels <-
c(
"dco2" = labels_breaks("dco2")$i_legend_title,
"dfco2" = labels_breaks("dfco2")$i_legend_title,
"atm_co2" = labels_breaks("atm_co2")$i_legend_title,
"atm_fco2" = labels_breaks("atm_fco2")$i_legend_title,
"sol" = labels_breaks("sol")$i_legend_title,
"kw" = labels_breaks("kw")$i_legend_title,
"kw_sol" = labels_breaks("kw_sol")$i_legend_title,
"intpp" = labels_breaks("intpp")$i_legend_title,
"no3" = labels_breaks("no3")$i_legend_title,
"o2" = labels_breaks("o2")$i_legend_title,
"dissic" = labels_breaks("dissic")$i_legend_title,
"sdissic" = labels_breaks("sdissic")$i_legend_title,
"cstar" = labels_breaks("cstar")$i_legend_title,
"talk" = labels_breaks("talk")$i_legend_title,
"stalk" = labels_breaks("stalk")$i_legend_title,
"sdissic_stalk" = labels_breaks("sdissic_stalk")$i_legend_title,
"spco2" = labels_breaks("spco2")$i_legend_title,
"sfco2" = labels_breaks("sfco2")$i_legend_title,
"sfco2_total" = labels_breaks("sfco2_total")$i_legend_title,
"sfco2_therm" = labels_breaks("sfco2_therm")$i_legend_title,
"sfco2_nontherm" = labels_breaks("sfco2_nontherm")$i_legend_title,
"fgco2" = labels_breaks("fgco2")$i_legend_title,
"fgco2_predict" = labels_breaks("fgco2_predict")$i_legend_title,
"fgco2_hov" = labels_breaks("fgco2_hov")$i_legend_title,
"fgco2_int" = labels_breaks("fgco2_int")$i_legend_title,
"fgco2_predict_int" = labels_breaks("fgco2_int")$i_legend_title,
"thetao" = labels_breaks("thetao")$i_legend_title,
"temperature" = labels_breaks("temperature")$i_legend_title,
"salinity" = labels_breaks("salinity")$i_legend_title,
"so" = labels_breaks("so")$i_legend_title,
"chl" = labels_breaks("chl")$i_legend_title,
"mld" = labels_breaks("mld")$i_legend_title,
"press" = labels_breaks("press")$i_legend_title,
"wind" = labels_breaks("wind")$i_legend_title,
"SSH" = labels_breaks("SSH")$i_legend_title,
"fice" = labels_breaks("fice")$i_legend_title,
"resid_fgco2" = labels_breaks("resid_fgco2")$i_legend_title,
"resid_fgco2_dfco2" = labels_breaks("resid_fgco2_dfco2")$i_legend_title,
"resid_fgco2_kw_sol" = labels_breaks("resid_fgco2_kw_sol")$i_legend_title,
"resid_fgco2_dfco2_kw_sol" = labels_breaks("resid_fgco2_dfco2_kw_sol")$i_legend_title,
"resid_fgco2_sum" = labels_breaks("resid_fgco2_sum")$i_legend_title,
"resid_fgco2_offset" = labels_breaks("resid_fgco2_offset")$i_legend_title
)Analysis settings
name_quadratic_fit <- c("atm_co2", "atm_fco2", "spco2", "sfco2")
# name_quadratic_fit <- c(name_quadratic_fit, "dfco2", "fgco2", "fgco2_int", "temperature")
start_year <- 1990
name_divergent <- c("dco2", "dfco2", "fgco2", "fgco2_hov", "fgco2_int")Data preprocessing
pco2_product <-
pco2_product %>%
filter(year >= start_year)pco2_product_interior <-
pco2_product_interior %>%
filter(time >= ymd(paste0(start_year, "-01-01")))biome_mask <- biome_mask %>%
mutate(area = earth_surf(lat, lon))
pco2_product <-
full_join(pco2_product,
biome_mask)
# set all values outside biome mask to NA
pco2_product <-
pco2_product %>%
mutate(across(-c(lat, lon, time, area, year, month, biome),
~ if_else(is.na(biome), NA, .)))Compuations
Maps
Biome means
pco2_product_biome_monthly_global <-
pco2_product %>%
filter(!is.na(fgco2)) %>%
mutate(fgco2_int = fgco2) %>%
mutate(biome = case_when(str_detect(biome, "SO-SPSS|SO-ICE|Arctic") ~ "Polar",
TRUE ~ "Global non-polar")) %>%
filter(biome == "Global non-polar") %>%
select(-c(lon, lat, year, month)) %>%
group_by(time, biome) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_monthly_biome <-
pco2_product %>%
filter(!is.na(fgco2)) %>%
mutate(fgco2_int = fgco2) %>%
select(-c(lon, lat, year, month)) %>%
group_by(time, biome) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_monthly <-
bind_rows(pco2_product_biome_monthly_global,
pco2_product_biome_monthly_biome)
rm(
pco2_product_biome_monthly_global,
pco2_product_biome_monthly_biome
)
pco2_product_biome_monthly <-
pco2_product_biome_monthly %>%
filter(!is.na(biome))
pco2_product_biome_monthly <-
pco2_product_biome_monthly %>%
mutate(year = year(time),
month = month(time),
.after = time)
pco2_product_biome_monthly <-
pco2_product_biome_monthly %>%
pivot_longer(-c(time, year, month, biome))
pco2_product_biome_annual <-
pco2_product_biome_monthly %>%
group_by(year, biome, name) %>%
summarise(value = mean(value)) %>%
ungroup()Profiles
pco2_product_interior <-
left_join(
biome_mask,
pco2_product_interior
)
pco2_product_profiles <- pco2_product_interior %>%
fselect(-c(lat, lon)) %>%
fgroup_by(biome, depth, time) %>% {
add_vars(fgroup_vars(., "unique"),
fmean(.,
w = area,
keep.w = FALSE,
keep.group_vars = FALSE))
}
pco2_product_profiles <-
pco2_product_profiles %>%
mutate(
year = year(time),
month = month(time)
)
gc()Zonal mean sections
pco2_product_interior <-
left_join(
region_mask,
pco2_product_interior %>% select(-c(biome, area))
)
pco2_product_zonal_mean <- pco2_product_interior %>%
fselect(-c(lon)) %>%
fgroup_by(region, depth, lat, time) %>% {
add_vars(fgroup_vars(., "unique"),
fmean(.,
keep.group_vars = FALSE))
}
pco2_product_zonal_mean <-
pco2_product_zonal_mean %>%
mutate(
year = year(time),
month = month(time)
)
gc()
rm(pco2_product_interior)
gc()Absolute values
Hovmoeller plots
The following Hovmoeller plots show the value of each variable as provided through the pCO2 product. Hovmoeller plots are first presented as annual means, and than as monthly means.
Annual means
pco2_product_hovmoeller_annual <-
pco2_product %>%
mutate(fgco2_int = fgco2) %>%
select(-c(lon, time, month, biome)) %>%
group_by(year, lat) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup() %>%
rename(fgco2_hov = fgco2_int) %>%
filter(fgco2_hov != 0)
pco2_product_hovmoeller_annual <-
pco2_product_hovmoeller_annual %>%
pivot_longer(-c(year, lat)) %>%
drop_na()
# pco2_product_hovmoeller_annual %>%
# filter(!(name %in% name_divergent)) %>%
# group_split(name) %>%
# # tail(5) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, lat, fill = value)) +
# geom_raster() +
# scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
# theme(legend.title = element_markdown()) +
# coord_cartesian(expand = 0) +
# labs(title = "Annual means",
# y = "Latitude") +
# theme(axis.title.x = element_blank())
# )
#
# pco2_product_hovmoeller_annual %>%
# filter(name %in% name_divergent) %>%
# group_split(name) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, lat, fill = value)) +
# geom_raster() +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks(.x %>% distinct(name)),
# limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
# oob = squish
# ) +
# theme(legend.title = element_markdown()) +
# coord_cartesian(expand = 0) +
# labs(title = "Annual means",
# y = "Latitude") +
# theme(axis.title.x = element_blank())
# )Monthly means
pco2_product_hovmoeller_monthly <-
pco2_product %>%
mutate(fgco2_int = fgco2) %>%
select(-c(lon, time, biome)) %>%
group_by(year, month, lat) %>%
summarise(across(-c(fgco2_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(fgco2_int,
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup() %>%
rename(fgco2_hov = fgco2_int) %>%
filter(fgco2_hov != 0)
pco2_product_hovmoeller_monthly <-
pco2_product_hovmoeller_monthly %>%
pivot_longer(-c(year, month, lat)) %>%
drop_na()
pco2_product_hovmoeller_monthly <-
pco2_product_hovmoeller_monthly %>%
mutate(decimal = year + (month-1) / 12)
# pco2_product_hovmoeller_monthly %>%
# filter(!(name %in% name_divergent)) %>%
# group_split(name) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(decimal, lat, fill = value)) +
# geom_raster() +
# scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
# theme(legend.title = element_markdown()) +
# labs(title = "Monthly means",
# y = "Latitude") +
# coord_cartesian(expand = 0) +
# theme(axis.title.x = element_blank())
# )
#
# pco2_product_hovmoeller_monthly %>%
# filter(name %in% name_divergent) %>%
# group_split(name) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(decimal, lat, fill = value)) +
# geom_raster() +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks(.x %>% distinct(name)),
# limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
# oob = squish
# )+
# theme(legend.title = element_markdown()) +
# labs(title = "Monthly means",
# y = "Latitude") +
# coord_cartesian(expand = 0) +
# theme(axis.title.x = element_blank())
# )rm(pco2_product)
gc()pCO2productanalysis_2023 <-
knitr::knit_expand(
file = here::here("analysis/child/pCO2_product_analysis.Rmd"),
product_name = "fCO2-Residual",
year_anom = 2023
)2023 anomalies
Functions
Anomaly detection
For the detection of anomalies at any point in time and space, we fit regression models and compare the fitted to the actual value.
We use linear regression models for all parameters, except for , which are approximated with quadratic fits.
The regression models are fitted to all data since , except 2023.
anomaly_determination <- function(df,...) {
group_by <- quos(...)
# group_by <- quos(lon, lat)
# group_by <- quos(biome)
# df <- pco2_product_map_annual
# Linear regression models
df_lm <-
df %>%
filter(year != 2023,
!(name %in% name_quadratic_fit)) %>%
drop_na() %>%
nest(data = -c(name, !!!group_by)) %>%
mutate(fit = map(data, ~ flm(
formula = value ~ year, data = .x
)))
df_lm <-
left_join(
df_lm %>%
unnest_wider(fit) %>%
select(name, !!!group_by,
intercept = `(Intercept)`, slope = year) %>%
mutate(intercept = as.vector(intercept),
slope = as.vector(slope)),
df
) %>%
mutate(fit = intercept + year * slope) %>%
select(name, !!!group_by, year, fit, value) %>%
mutate(resid = value - fit)
# df_lm <-
# df %>%
# filter(year != 2023,
# !(name %in% name_quadratic_fit)) %>%
# drop_na() %>%
# nest(data = -c(name, !!!group_by)) %>%
# mutate(
# fit = map(data, ~ lm(value ~ year, data = .x)),
# tidied = map(fit, tidy),
# augmented = map(fit, augment)
# )
#
#
# df_lm_year_anom <-
# full_join(
# df_lm %>%
# unnest(tidied) %>%
# select(name, !!!group_by, term, estimate) %>%
# pivot_wider(names_from = term,
# values_from = estimate) %>%
# mutate(fit = `(Intercept)` + year * 2023) %>%
# select(name, !!!group_by, fit) %>%
# mutate(year = 2023),
# df %>%
# filter(year == 2023,
# !(name %in% name_quadratic_fit))
# ) %>%
# mutate(resid = value - fit)
#
#
# df_lm <-
# bind_rows(
# df_lm %>%
# unnest(augmented) %>%
# select(name, !!!group_by, year, value, fit = .fitted, resid = .resid),
# df_lm_year_anom
# )
#
# rm(df_lm_year_anom)
# Quadratic regression models
if(any(df %>% distinct(name) %>% pull() %in% name_quadratic_fit)){
df_quadratic <-
df %>%
filter(year != 2023,
name %in% name_quadratic_fit) %>%
drop_na() %>%
mutate(year = year - 2000) %>%
nest(data = -c(name, !!!group_by)) %>%
mutate(
fit = map(data, ~ flm(
# formula = value ~ year + I(year ^ 2), data = .x))
# formula = value ~ year + I(year ^ 2) + I(year ^ 3), data = .x))
formula = value ~ poly(year, 2, raw = TRUE), data = .x))
)
df_quadratic <-
left_join(
df_quadratic %>%
unnest_wider(fit) %>%
select(name, !!!group_by,
intercept = `(Intercept)`,
slope = `1`,
slope_squared = `2`
# slope_cube = `3`
) %>%
mutate(intercept = as.vector(intercept),
slope = as.vector(slope),
slope_squared = as.vector(slope_squared)
# slope_cube = as.vector(slope_cube)
),
df %>%
mutate(year = year - 2000)
) %>%
mutate(fit = intercept + year * slope + year^2 * slope_squared) %>%
select(name, !!!group_by, year, fit, value) %>%
mutate(resid = value - fit,
year = year + 2000)
# df_quadratic <-
# df %>%
# filter(year != 2023,
# name %in% name_quadratic_fit) %>%
# nest(data = -c(name, !!!group_by)) %>%
# mutate(
# fit = map(data, ~ lm(value ~ year + I(year ^ 2), data = .x)),
# tidied = map(fit, tidy),
# augmented = map(fit, augment)
# )
#
# df_quadratic_year_anom <-
# full_join(
# df_quadratic %>%
# unnest(tidied) %>%
# select(name, !!!group_by, term, estimate) %>%
# pivot_wider(names_from = term,
# values_from = estimate) %>%
# mutate(fit = `(Intercept)` + year * 2023 + `I(year^2)` * 2023 ^ 2) %>%
# select(name, !!!group_by, fit) %>%
# mutate(year = 2023),
# df %>%
# filter(year == 2023,
# name %in% name_quadratic_fit)
# ) %>%
# mutate(resid = value - fit)
#
#
# df_quadratic <-
# bind_rows(
# df_quadratic %>%
# unnest(augmented) %>%
# select(name, !!!group_by, year, value, fit = .fitted, resid = .resid),
# df_quadratic_year_anom
# )
#
# rm(df_quadratic_year_anom)
# Join linear and quadratic regression results
df_anomaly <-
bind_rows(df_lm,
df_quadratic)
rm(df_lm,
df_quadratic)
} else{
df_anomaly <- df_lm
rm(df_lm)
}
df_anomaly <-
df_anomaly %>%
arrange(year)
return(df_anomaly)
}Seasonality plots
warm_color <- "#B84A60FF"
cold_color <- "#16877CFF"
p_season <- function(df,
dim_row = "name",
dim_col = "biome",
title = NULL,
var = "resid",
scales = "free_y") {
p <- ggplot(data = df,
aes(month, !!ensym(var)))
if(var == "resid"){
p <- p +
geom_hline(yintercept = 0, linewidth =0.5)
}
p <- p +
geom_path(data = . %>% filter(year != 2023),
aes(group = as.factor(year),
col = as.factor(paste(min(year), max(year), sep = "-"))),
alpha = 0.5)+
geom_path(data = . %>%
filter(year != 2023) %>%
group_by_at(vars(month, dim_col, dim_row)) %>%
summarise(!!ensym(var) := mean(!!ensym(var))),
aes(col = "Climatological\nmean"),
linewidth = 1) +
scale_color_manual(values = c("grey", "black"),
guide = guide_legend(order = 2,
reverse = TRUE)) +
new_scale_color()+
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(
values = warm_color,
guide = guide_legend(order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = title,
x = "Month")
if(df %>% filter(name == "fgco2") %>% nrow() > 0 & "value" %in% names(df)){
df_sink <- df %>%
filter(year == 2023,
name == "fgco2")
p <- p +
geom_point(data = df_sink %>% filter(value < 0),
aes(shape = "Sink"), fill = "white") +
geom_point(data = df_sink %>% filter(value >= 0),
aes(shape = "Source"), fill = "white") +
scale_shape_manual(values = c(25,24))
}
if (!(is.null(dim_col))) {
p <- p +
facet_grid(
as.formula(paste(dim_row, "~", dim_col)),
scales = scales,
labeller = labeller(name = x_axis_labels),
switch = "y"
)
} else {
p <- p +
facet_grid(
as.formula(paste(dim_row, "~ .")),
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
)
}
p <- p +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)
p
}fCO2 decomposition
fco2_decomposition <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
# group_by <- quos(biome, year, month)
pco2_product_biome_monthly_fCO2_decomposition <-
df %>%
filter(name %in% c("temperature", "sfco2"))
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "temperature") %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid),
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "sfco2") %>%
select(-c(value, resid)) %>%
pivot_wider(values_from = fit)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(sfco2_therm = (sfco2 * exp(0.0423 * temperature)) - sfco2)
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition,
df %>%
filter(name %in% c("sfco2")) %>%
select(-c(value, fit, name)) %>%
rename(sfco2_total = resid)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(sfco2_nontherm = sfco2_total - sfco2_therm)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
select(-c(temperature, sfco2)) %>%
pivot_longer(starts_with("sfco2"),
values_to = "resid")
}fco2_decomposition <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
# group_by <- quos(biome, year, month)
pco2_product_biome_monthly_fCO2_decomposition <-
df %>%
filter(name %in% c("temperature", "sfco2"))
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "temperature") %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid),
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(name == "sfco2") %>%
select(-c(value, resid)) %>%
pivot_wider(values_from = fit)
)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
mutate(
sfco2_therm = (sfco2 * exp(0.0423 * temperature)) - sfco2,
sfco2_nontherm = (sfco2 * exp(-0.0423 * temperature)) - sfco2)
pco2_product_biome_monthly_fCO2_decomposition <-
inner_join(
pco2_product_biome_monthly_fCO2_decomposition,
df %>%
filter(name %in% c("sfco2")) %>%
select(-c(value, fit, name)) %>%
rename(sfco2_total = resid)
)
# pco2_product_biome_monthly_fCO2_decomposition <-
# pco2_product_biome_monthly_fCO2_decomposition %>%
# mutate(sfco2_nontherm = sfco2_total - sfco2_therm)
pco2_product_biome_monthly_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
select(-c(temperature, sfco2)) %>%
pivot_longer(starts_with("sfco2"),
values_to = "resid")
}Flux attribution
flux_attribution <- function(df, ...) {
group_by <- quos(...)
# group_by <- quos(lon, lat, month)
pco2_product_flux_attribution <-
df %>%
filter(name %in% c("dfco2", "kw_sol", "fgco2"))
pco2_product_flux_attribution <-
inner_join(
pco2_product_flux_attribution %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid,
names_prefix = "resid_"),
pco2_product_flux_attribution %>%
select(-c(value, resid)) %>%
filter(name != "fgco2") %>%
pivot_wider(values_from = fit)
)
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
mutate(
resid_fgco2_dfco2 = resid_dfco2 * kw_sol,
resid_fgco2_kw_sol = resid_kw_sol * dfco2,
resid_fgco2_dfco2_kw_sol = resid_dfco2 * resid_kw_sol
# resid_fgco2_sum = resid_fgco2_dfco2 + resid_fgco2_kw_sol + resid_fgco2_dfco2_kw_sol
)
# pco2_product_flux_attribution <-
# pco2_product_flux_attribution %>%
# mutate(resid_fgco2_offset = resid_fgco2 - resid_fgco2_sum)
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
select(!!!group_by, starts_with("resid_fgco2")) %>%
pivot_longer(starts_with("resid_"),
values_to = "resid")
pco2_product_flux_attribution <-
pco2_product_flux_attribution %>%
filter(str_detect(name, "dfco2|kw_sol")) %>%
mutate(name = factor(
name,
levels = c(
"resid_fgco2",
"resid_fgco2_dfco2",
"resid_fgco2_kw_sol",
"resid_fgco2_dfco2_kw_sol",
"resid_fgco2_sum",
"resid_fgco2_offset"
)
))
}Maps
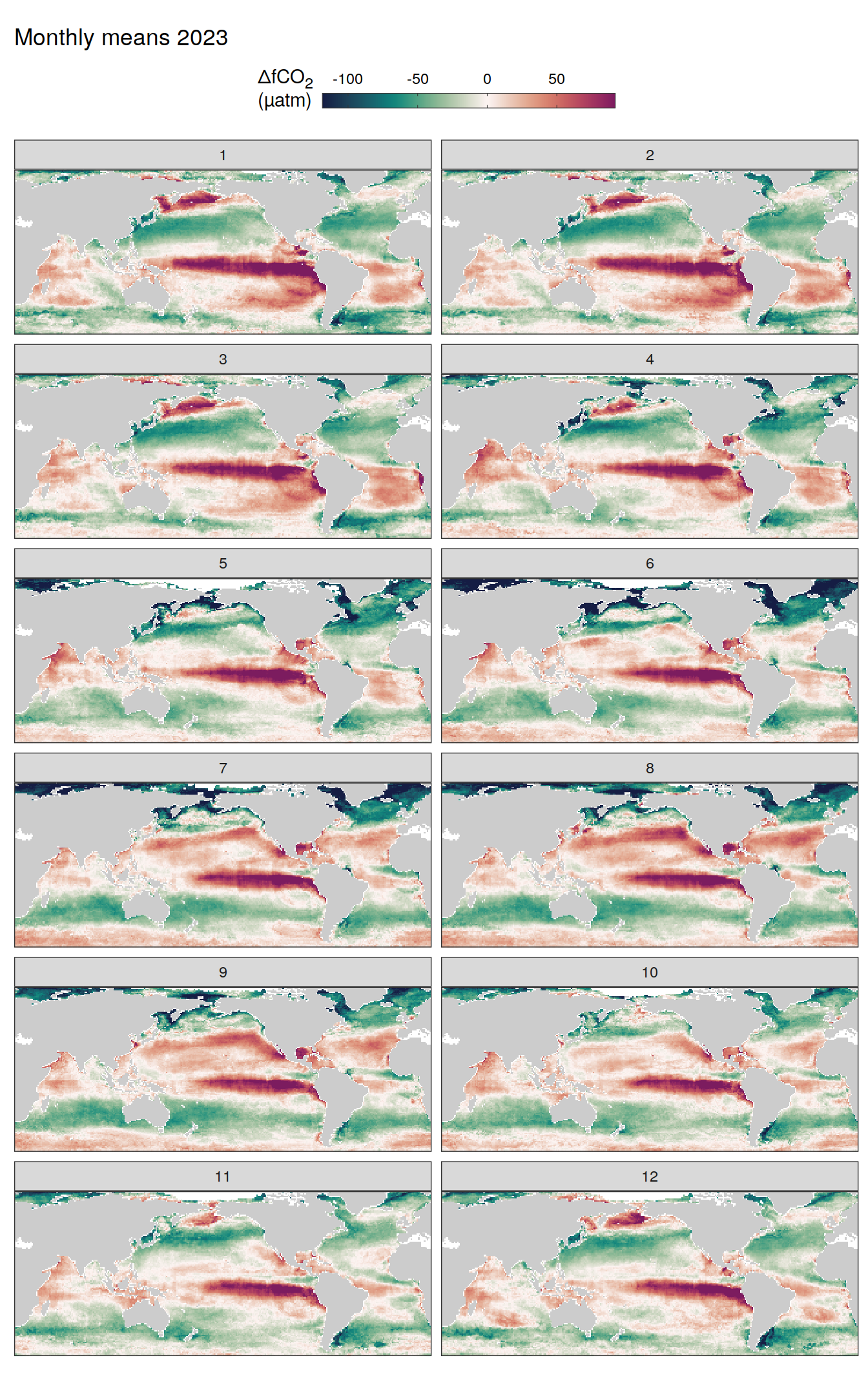
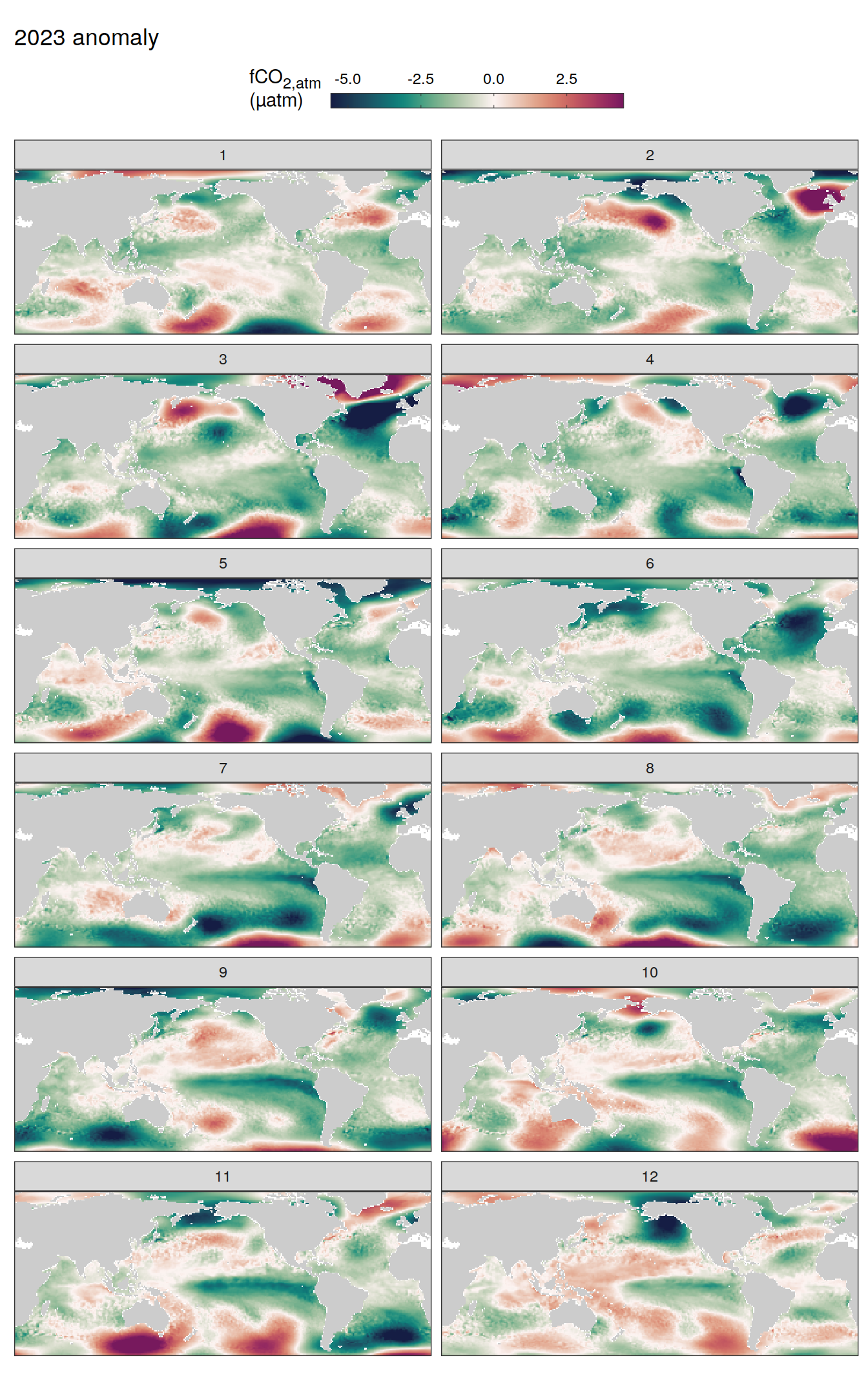
The following maps show the absolute state of each variable in 2023 as provided through the pCO2 product, the change in that variable from 1990 to 2023, as well es the anomalies in 2023. Changes and anomalies are determined based on the predicted value of a linear regression model fit to the data from 1990 to 2022.
Maps are first presented as annual means, and than as monthly means. Note that the 2023 predictions for the monthly maps are done individually for each month, such the mean seasonal anomaly from the annual mean is removed.
Note: The increase the computational speed, I regridded all maps to 5X5° grid.
Annual means
2023 absolute
pco2_product_map_annual_anomaly <-
pco2_product_map_annual %>%
drop_na() %>%
anomaly_determination(lon, lat)
pco2_product_map_annual_anomaly <-
pco2_product_map_annual_anomaly %>%
drop_na()
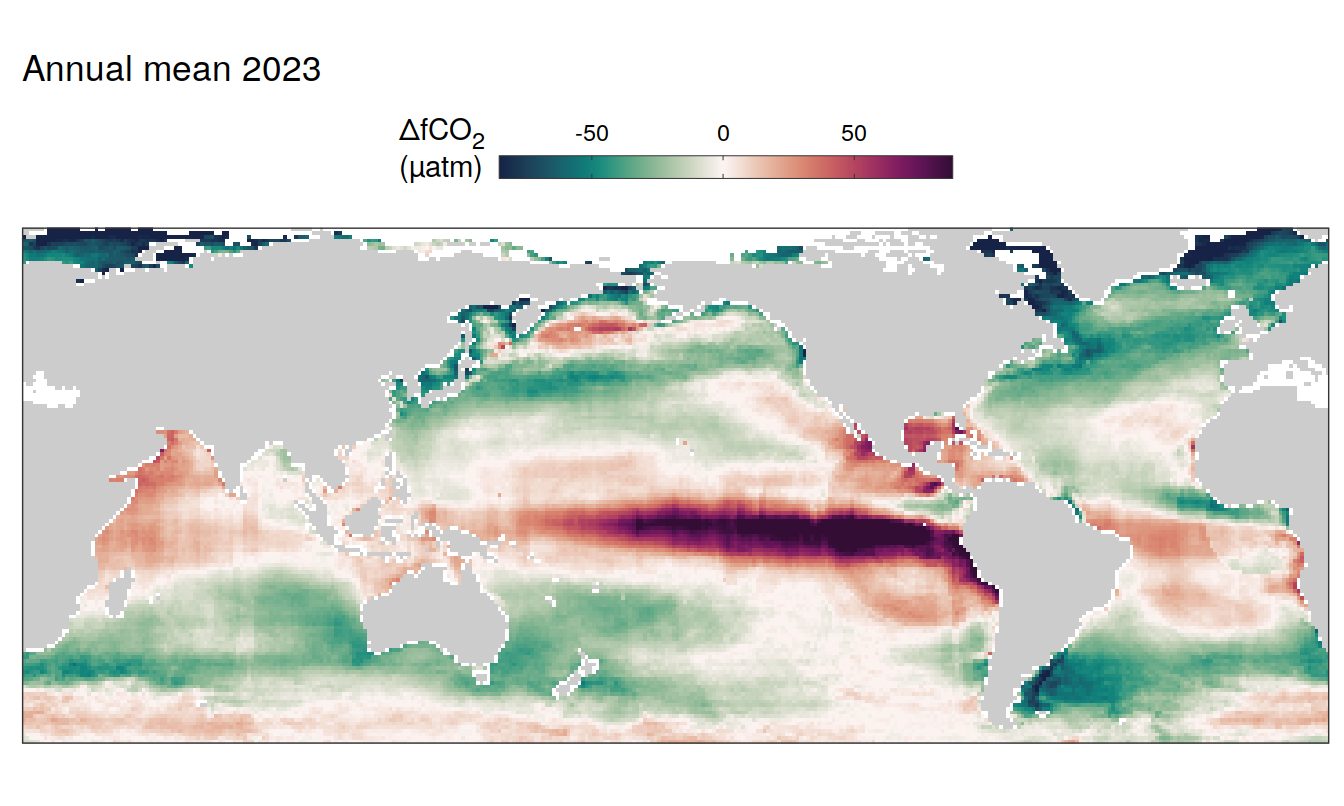
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
!(name %in% name_divergent)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = value)) +
labs(title = paste("Annual mean", 2023)) +
scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)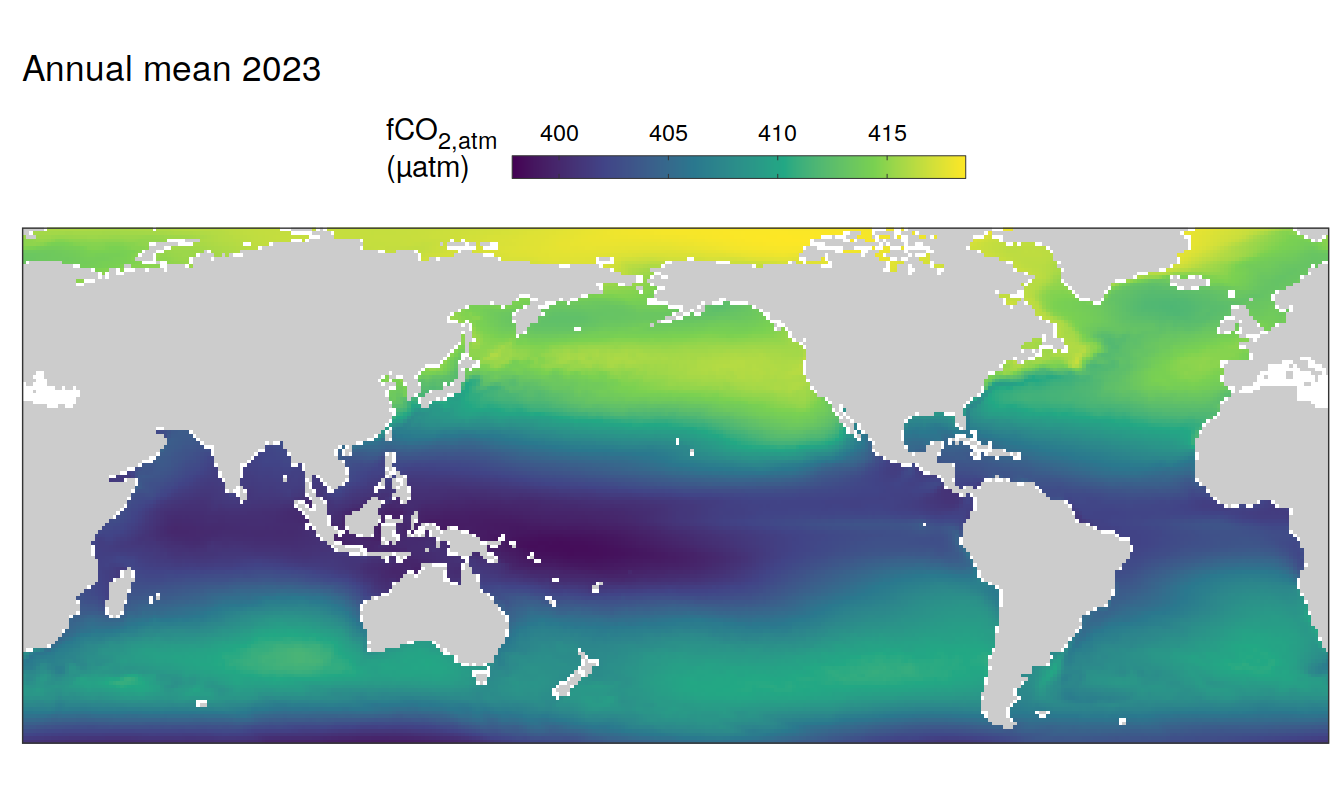
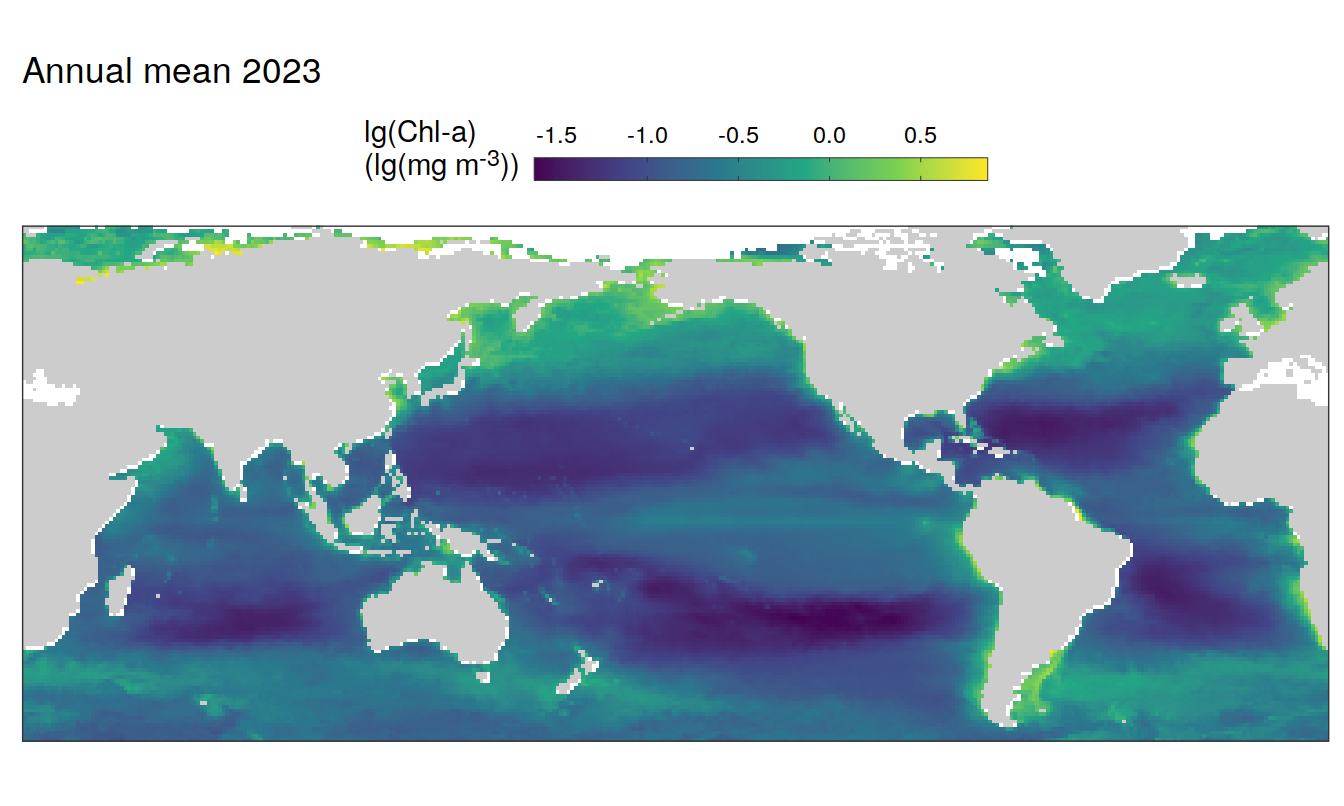


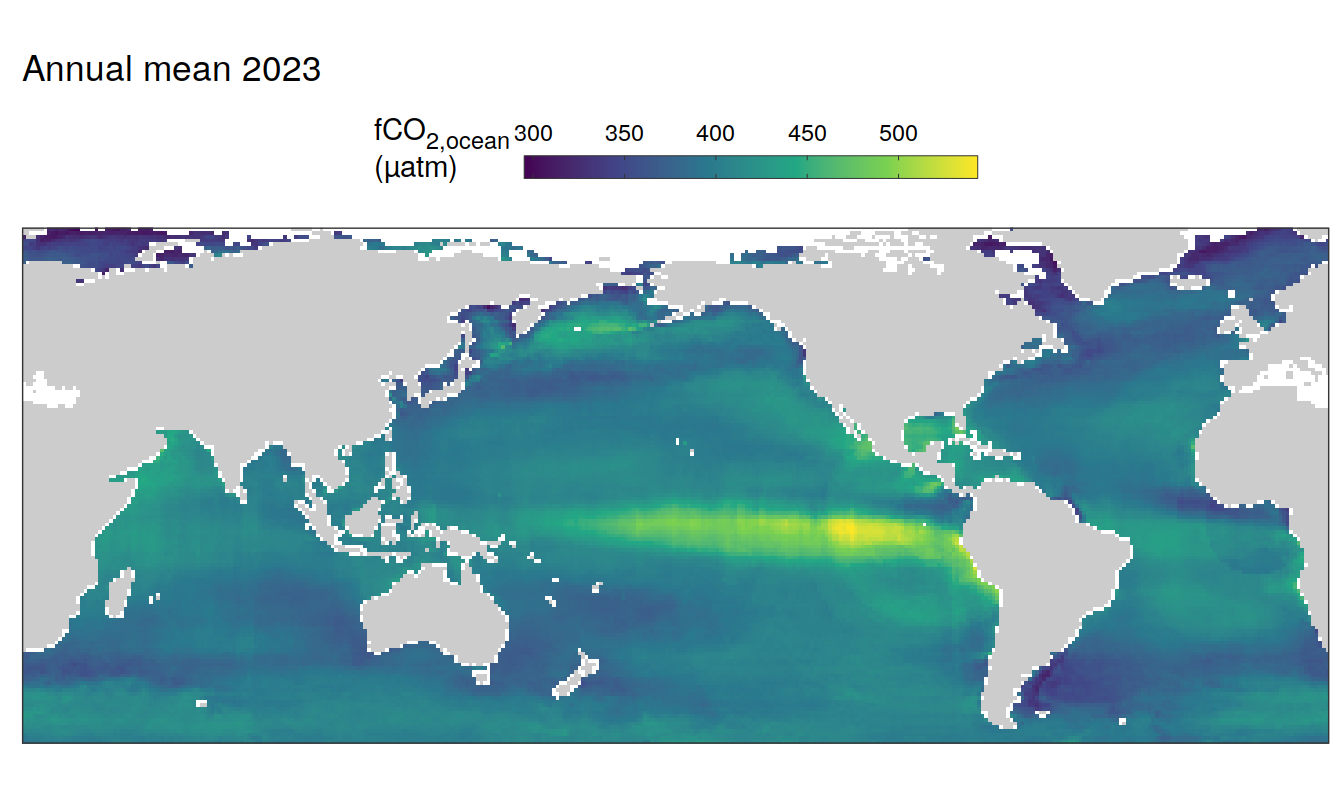
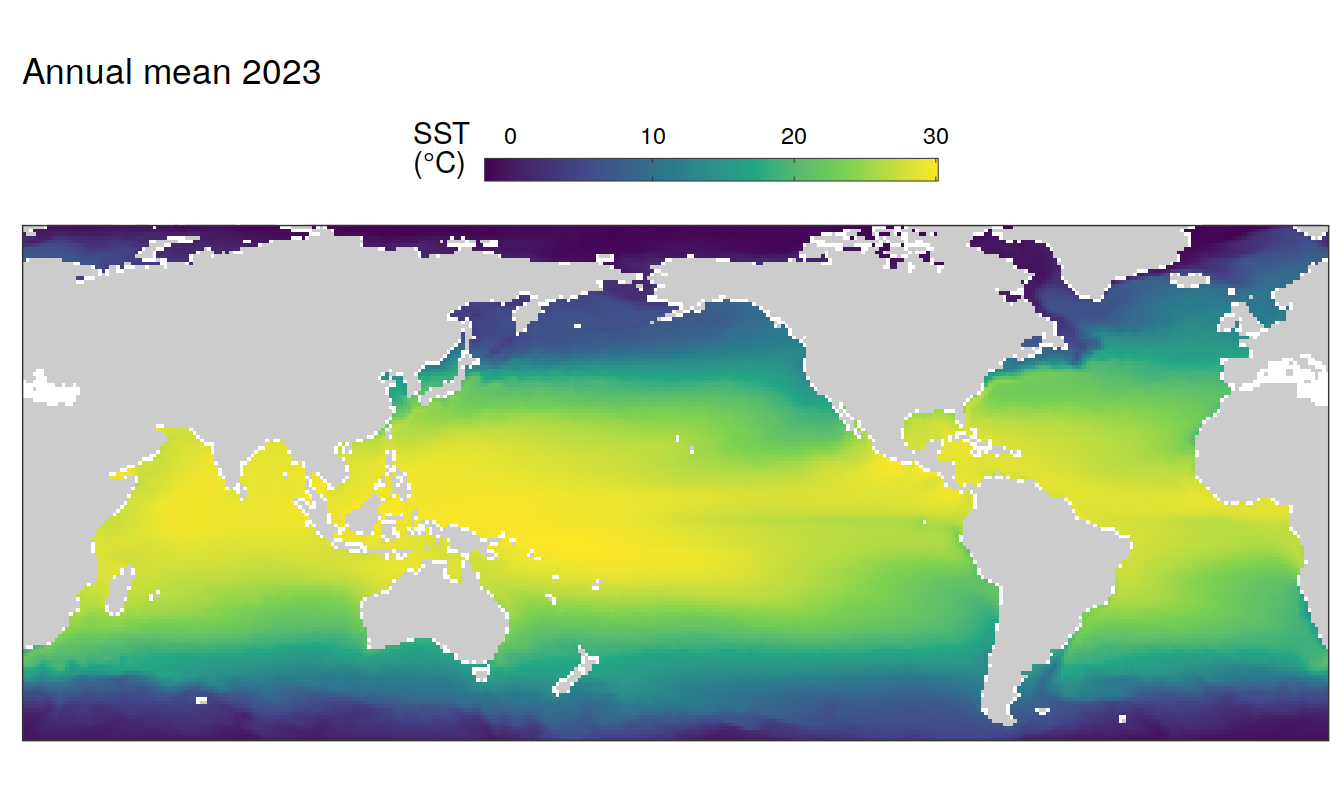
pco2_product_map_annual_anomaly %>%
filter(year == 2023,
name %in% name_divergent) %>%
group_split(name) %>%
# head(1) %>%
map( ~ map +
geom_tile(data = .x,
aes(lon, lat, fill = value)) +
labs(title = paste("Annual mean", 2023)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)
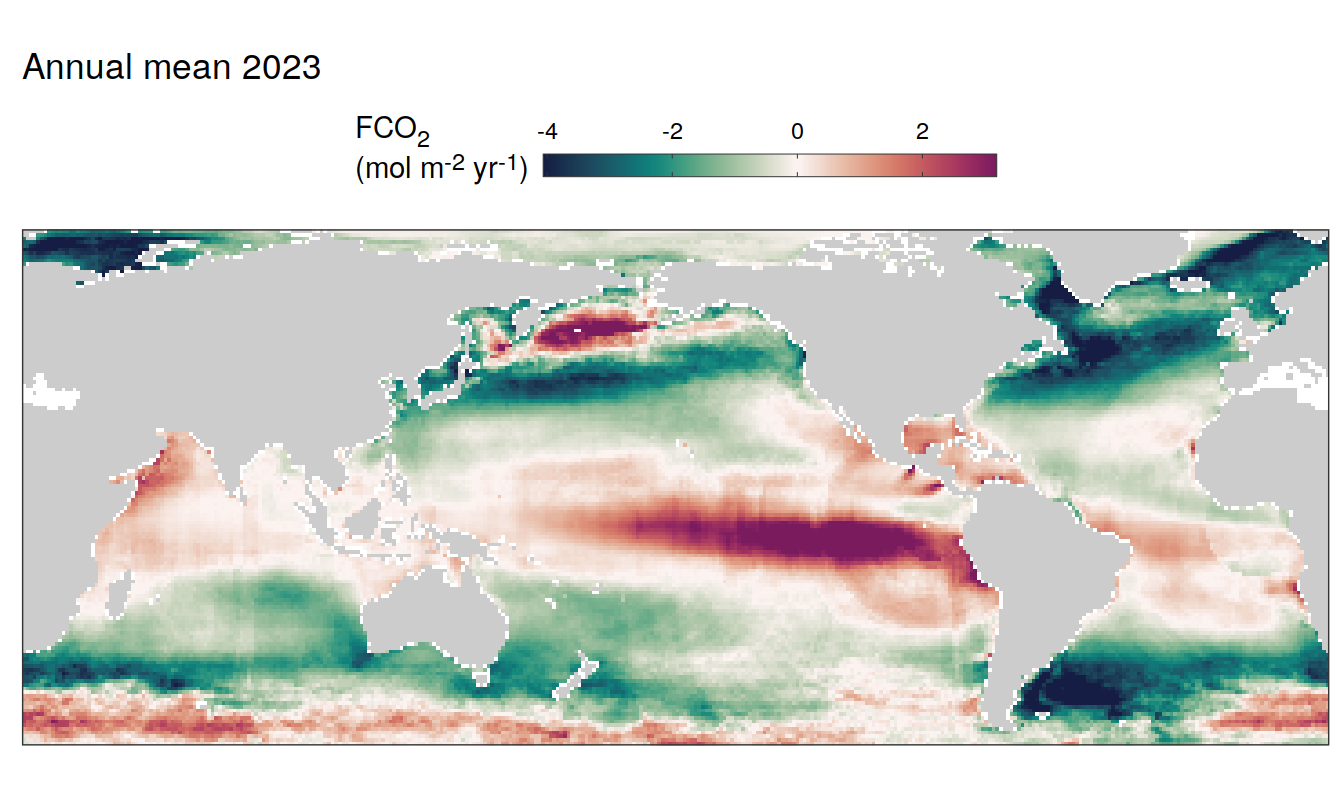
Trends
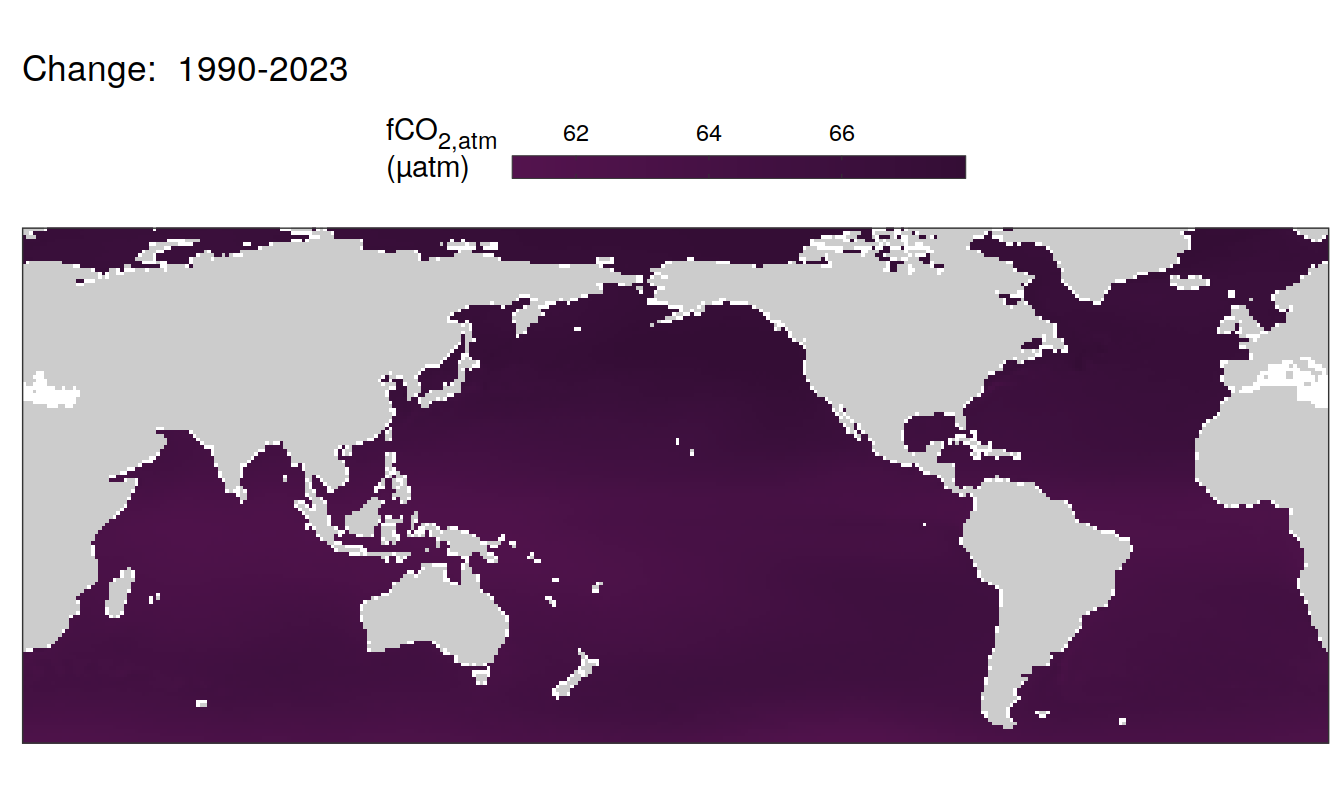
pco2_product_map_annual_anomaly %>%
group_by(name) %>%
filter(year %in% c(min(year), max(year), 2023)) %>%
ungroup() %>%
select(-c(value, resid)) %>%
filter(year %in% c(min(year), max(year))) %>%
arrange(year) %>%
group_by(lon, lat, name) %>%
mutate(change = fit - lag(fit),
period = paste(lag(year), year, sep = "-")) %>%
ungroup() %>%
filter(!is.na(change)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(lon, lat, fill = change)) +
labs(title = paste("Change: ",.x$period)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$change, .01), quantile(.x$change, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)
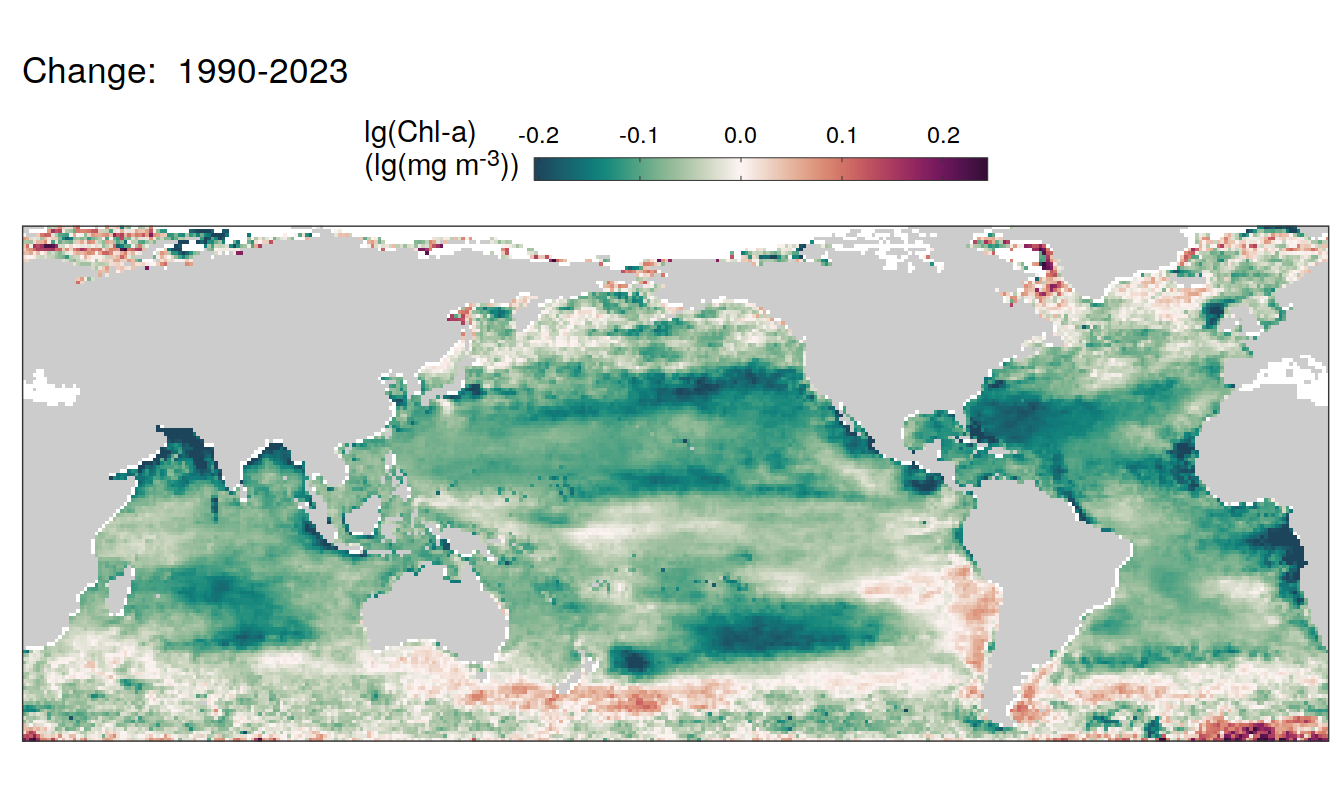
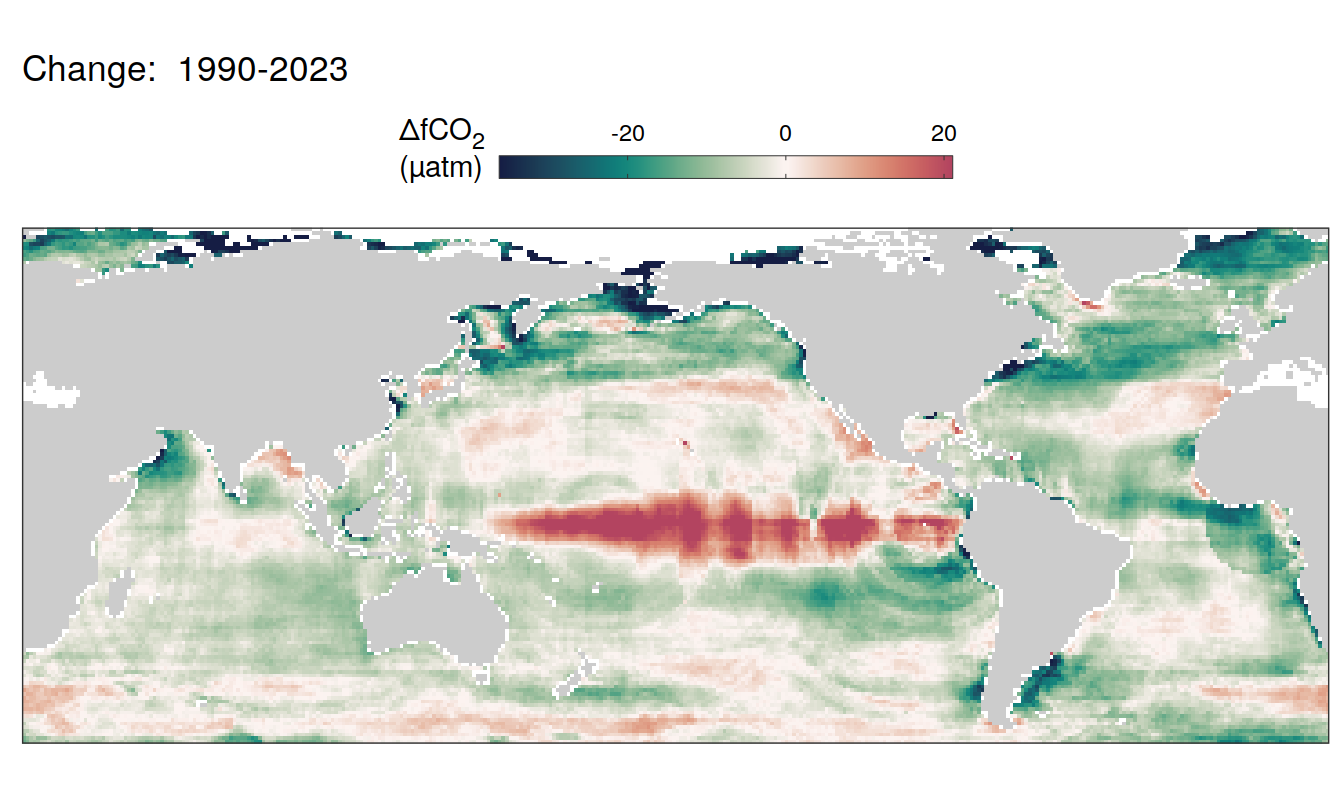
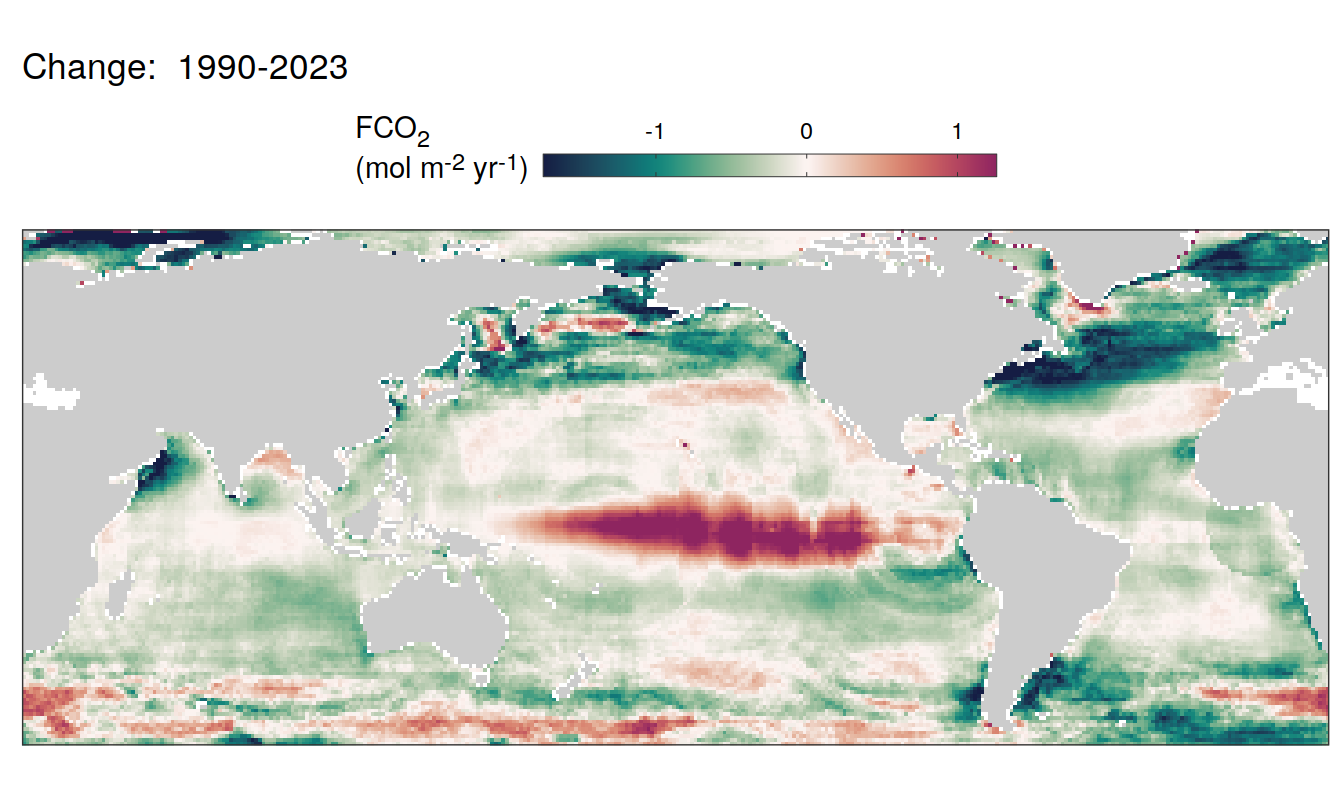
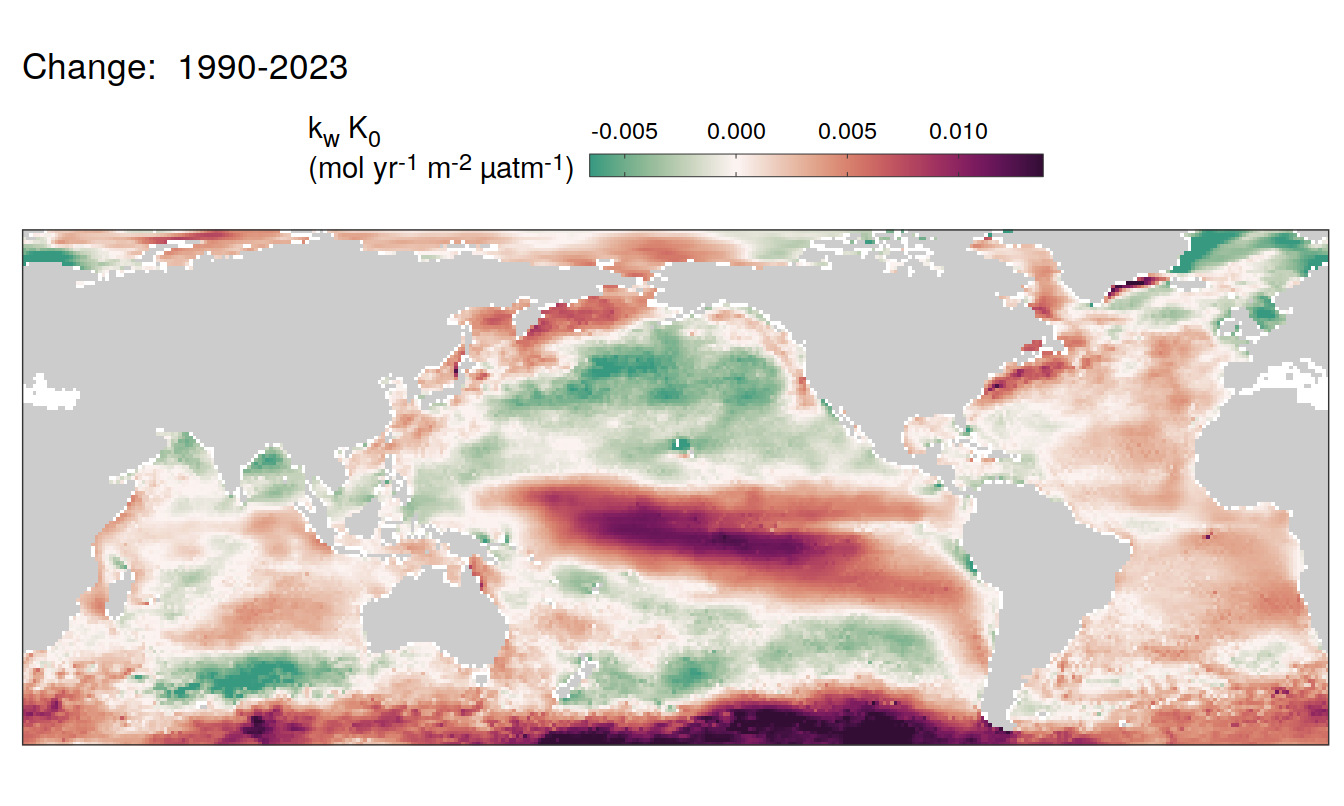
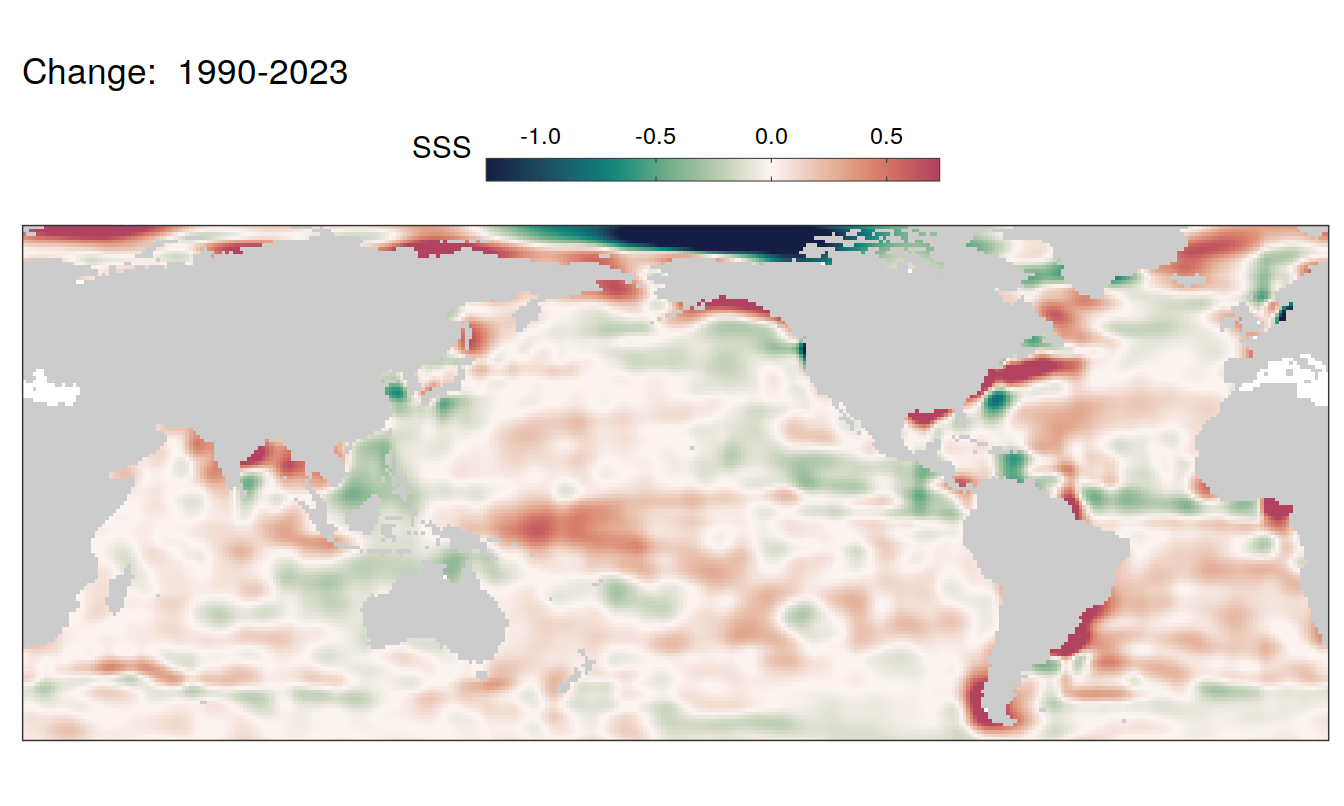
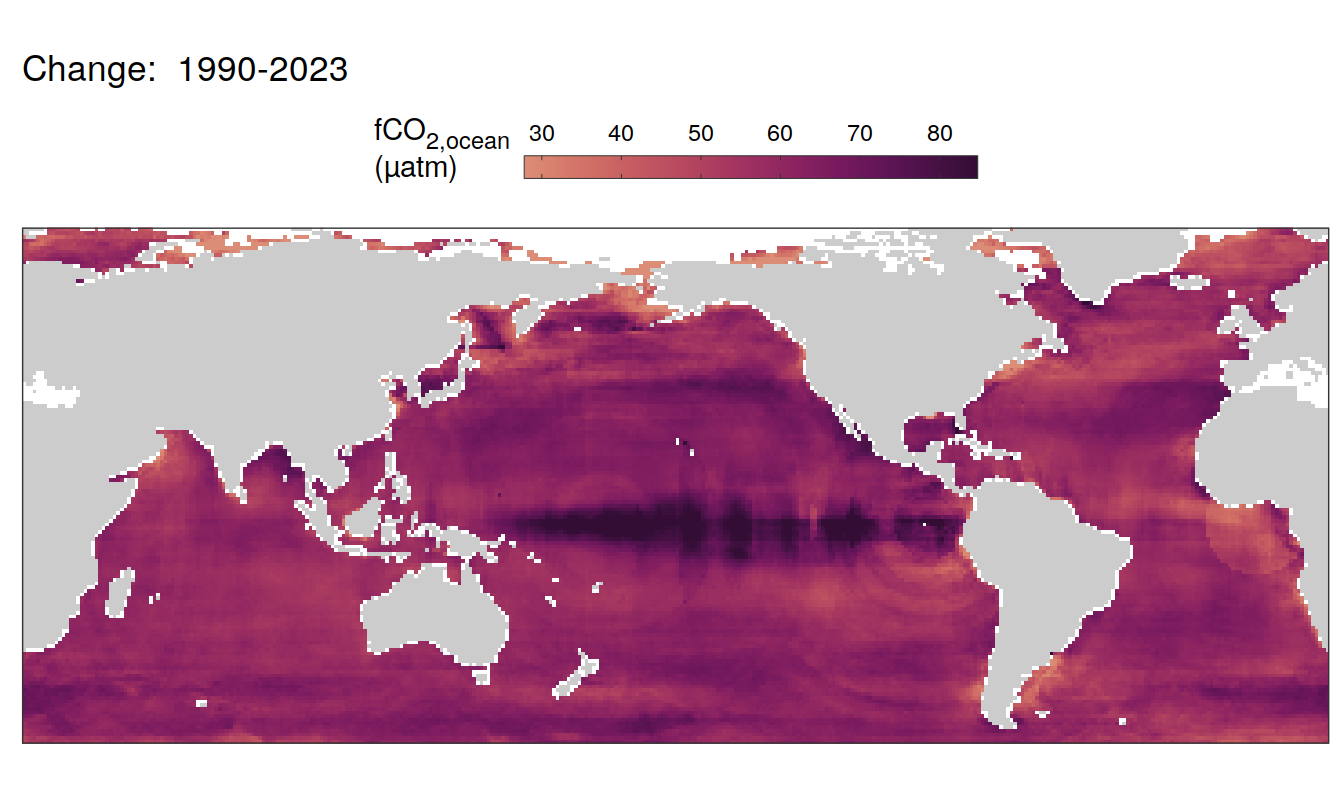
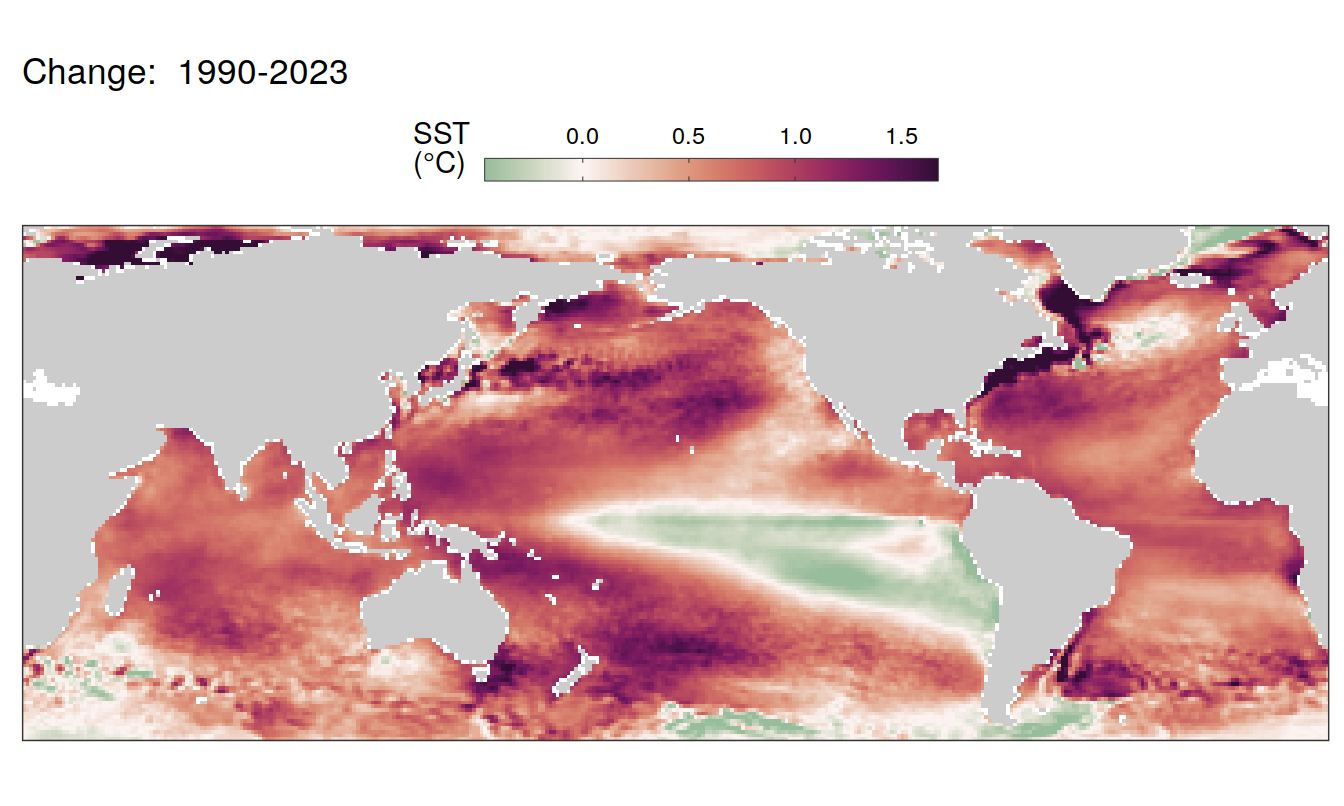
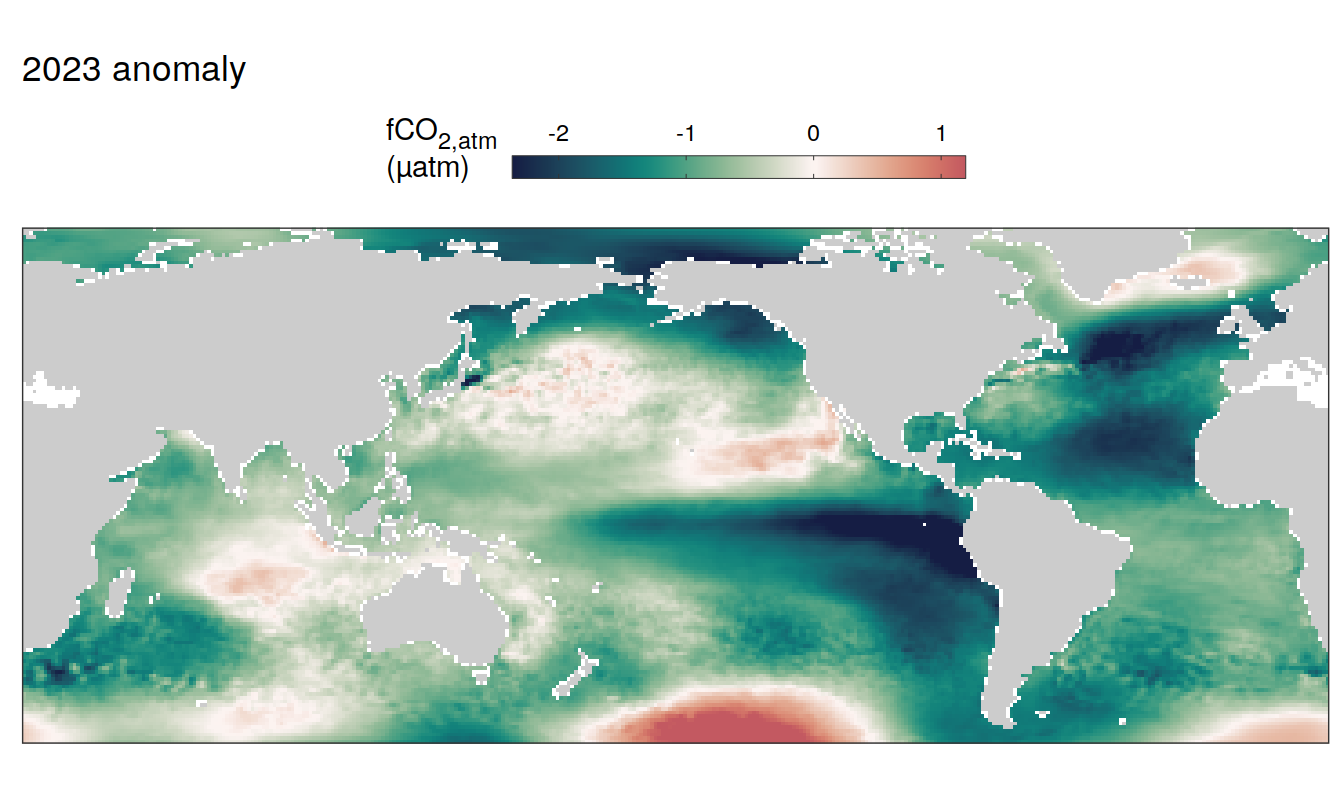
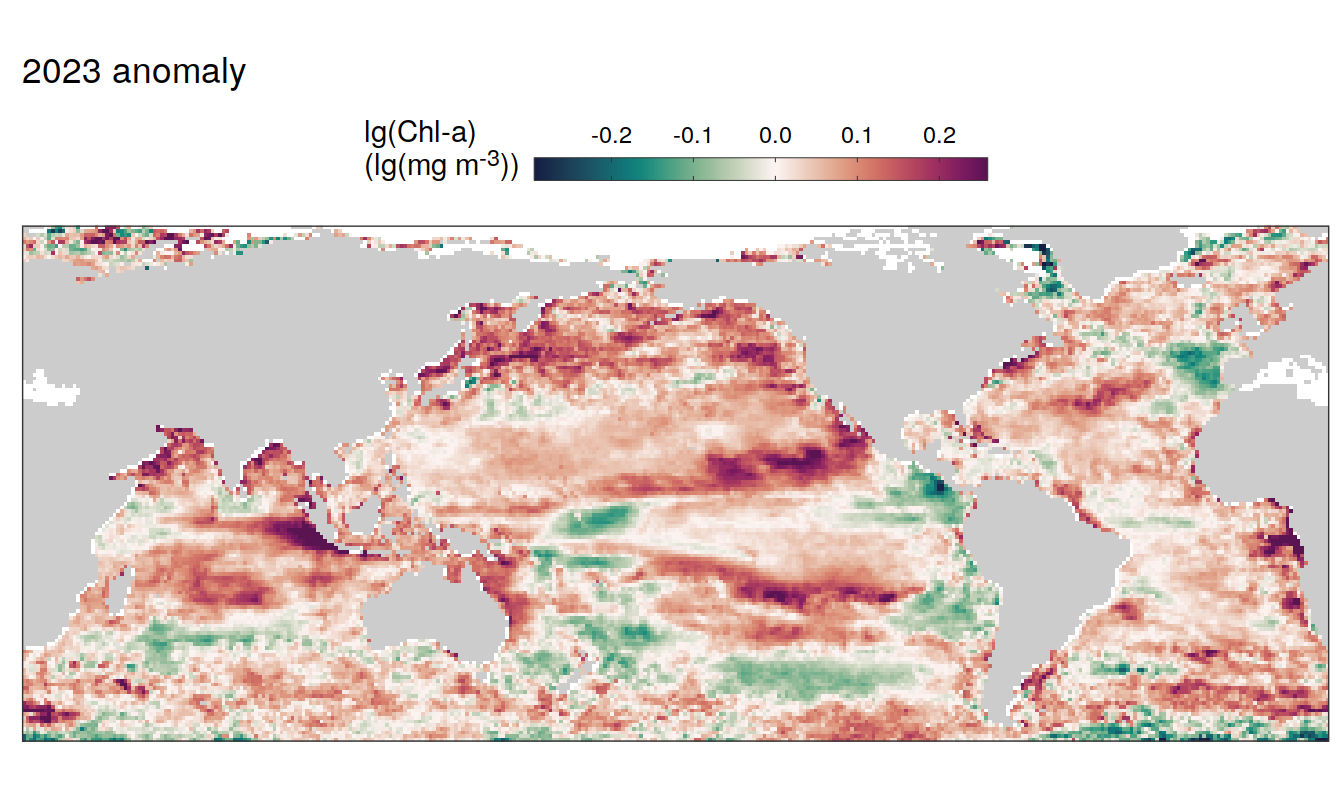
2023 anomaly
pco2_product_map_annual_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(1) %>%
map( ~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid)) +
labs(title = paste(2023,"anomaly")) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
)+
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)
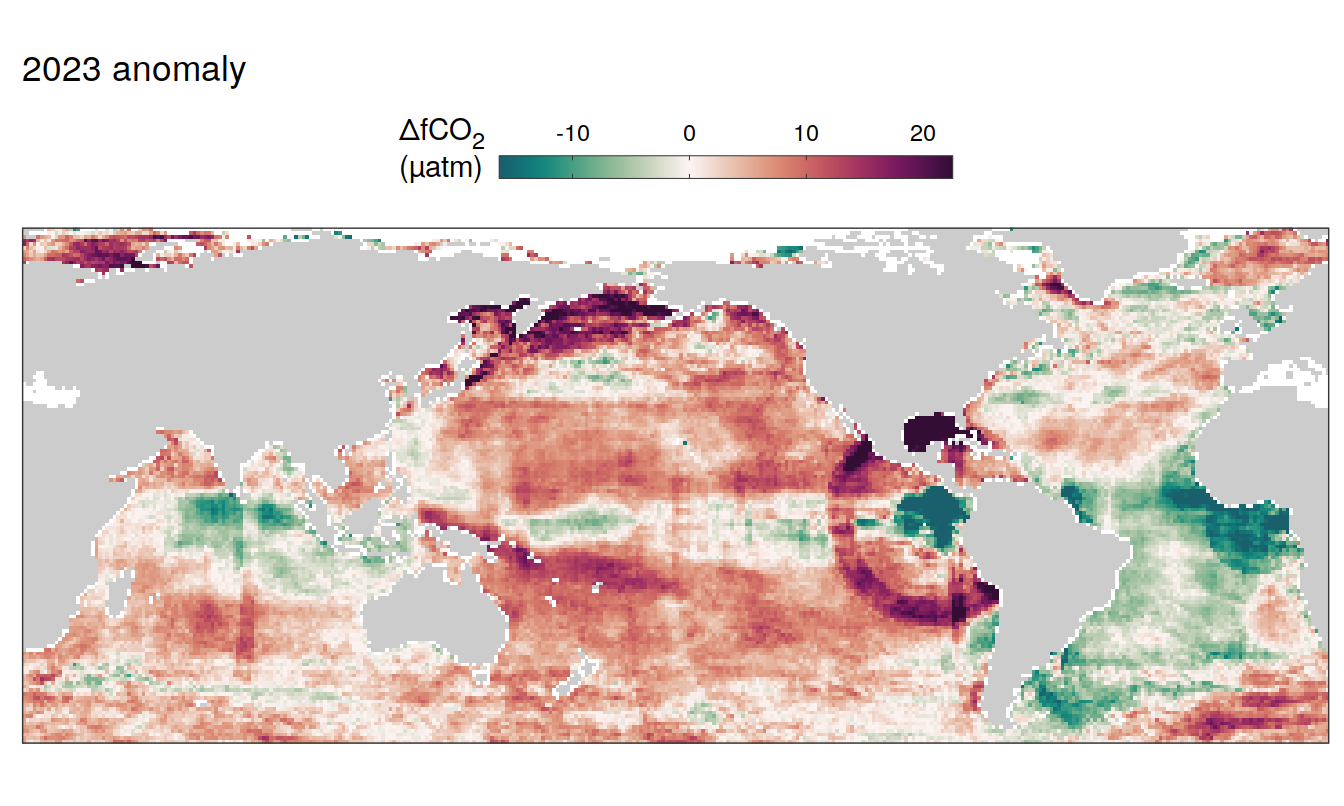
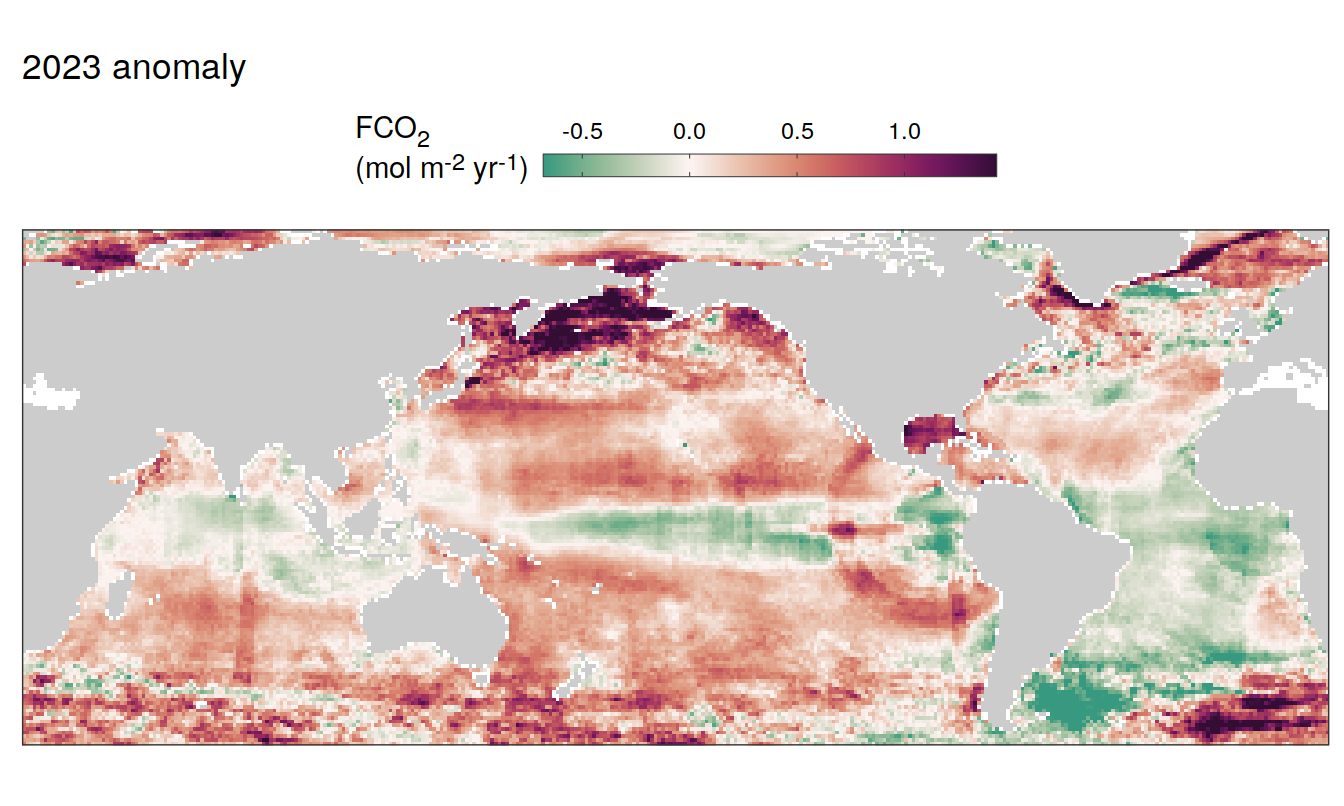
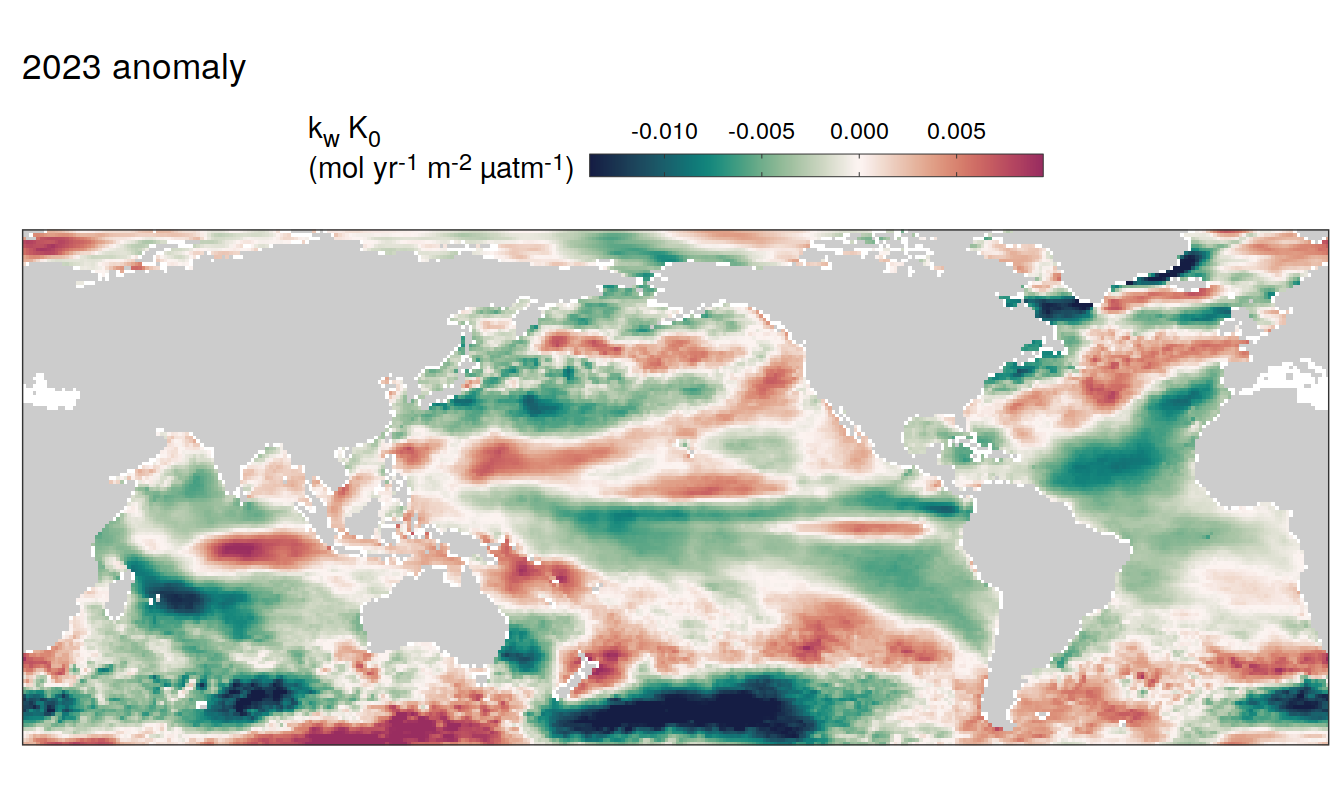
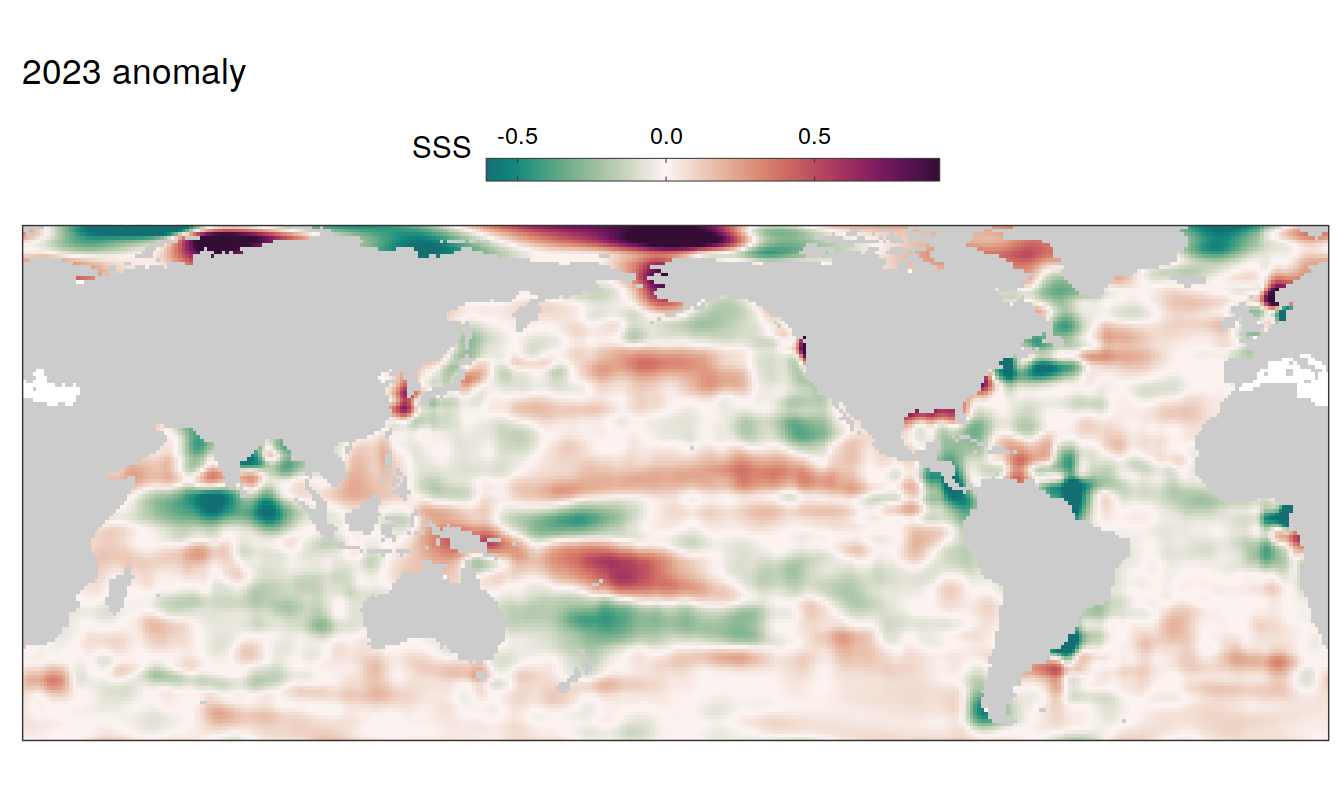
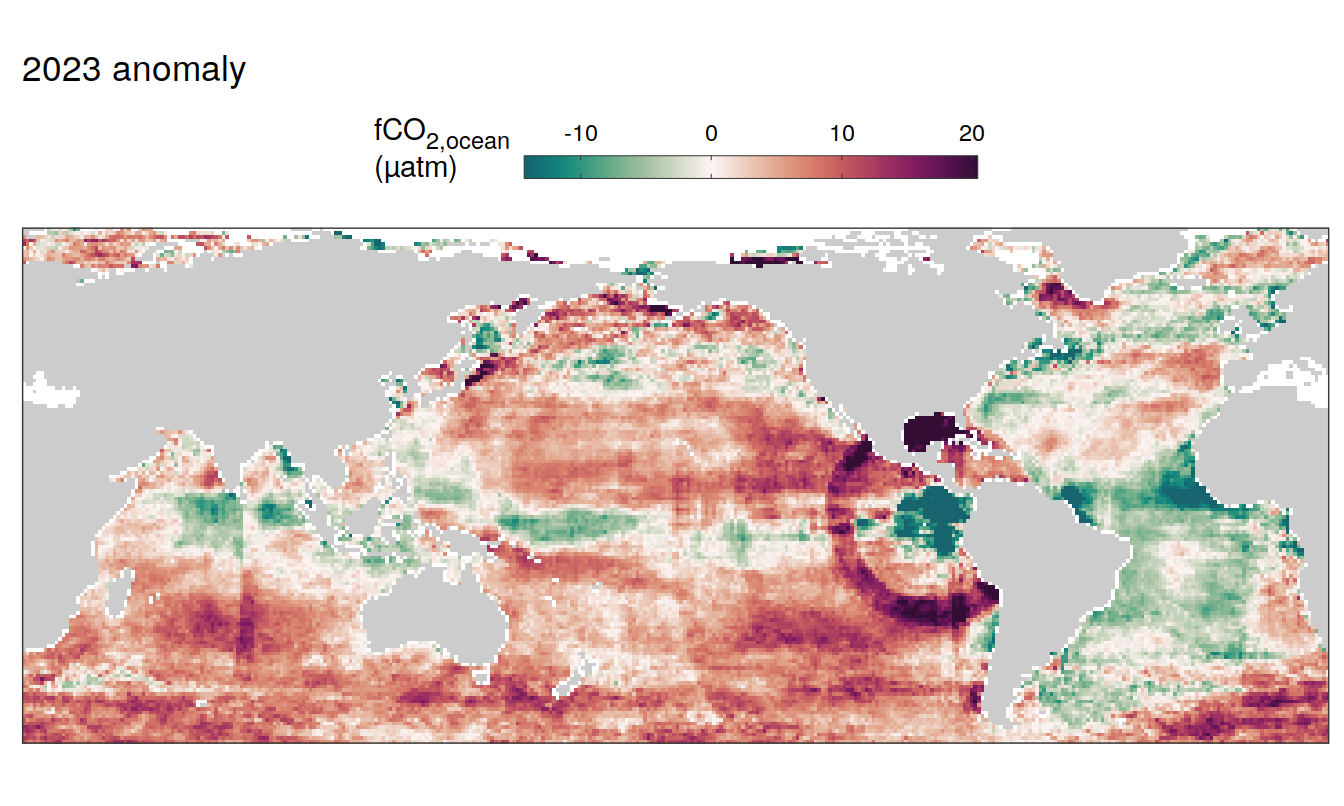
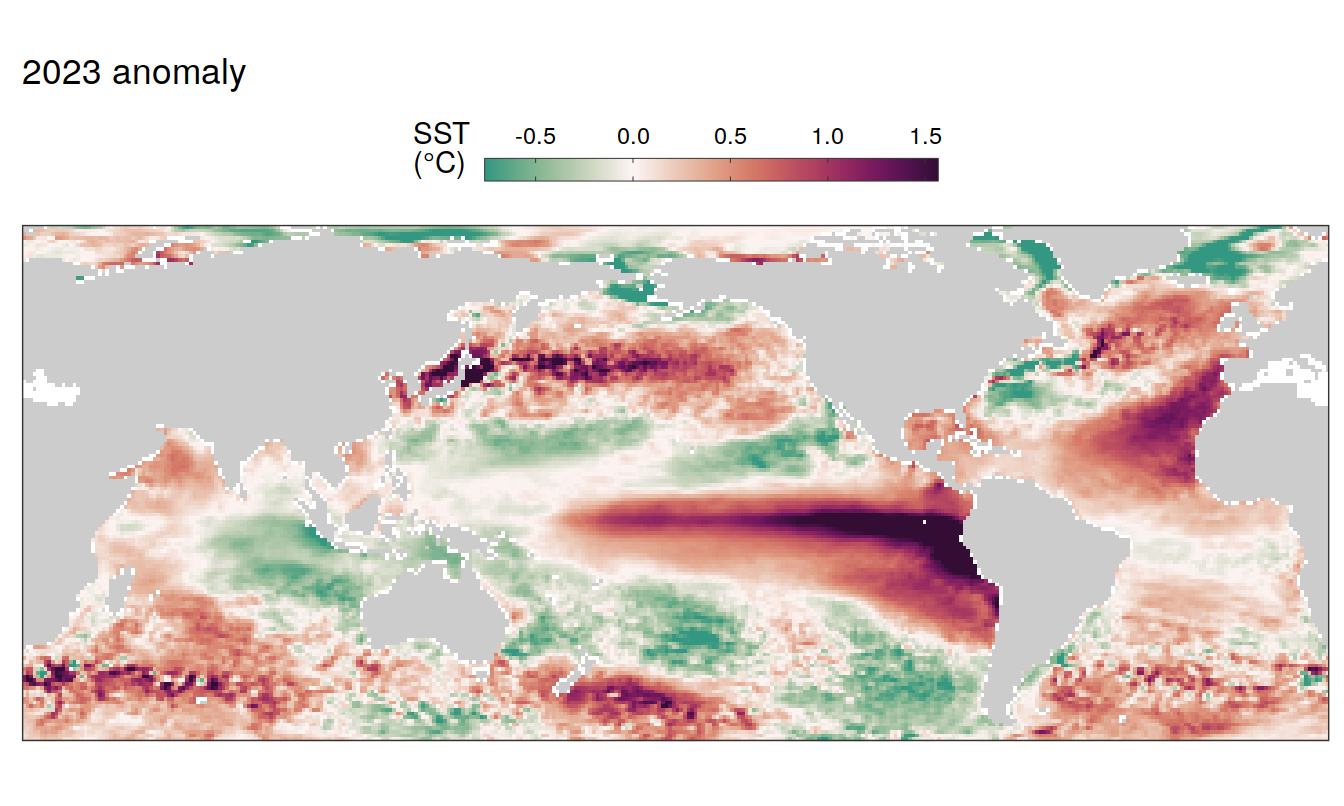
SST flux slope
pco2_product_map_annual_slope <-
pco2_product_map_annual_anomaly %>%
filter(year != 2023) %>%
select(year, lon, lat, resid, name) %>%
pivot_wider(values_from = resid) %>%
select(lon, lat, fgco2, temperature) %>%
drop_na() %>%
nest(data = -c(lon, lat)) %>%
mutate(fit = map(data, ~ flm(
formula = fgco2 ~ temperature, data = .x
)))
pco2_product_map_annual_slope <-
pco2_product_map_annual_slope %>%
unnest_wider(fit) %>%
select(lon, lat, slope = temperature) %>%
mutate(slope = as.vector(slope))
map +
geom_tile(data = pco2_product_map_annual_slope,
aes(lon, lat, fill = slope)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
limits = c(
quantile(pco2_product_map_annual_slope$slope,.01),
quantile(pco2_product_map_annual_slope$slope, .99)),
oob = squish
) +
labs(title = "Correlation of historic annual flux and SST anomalies") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")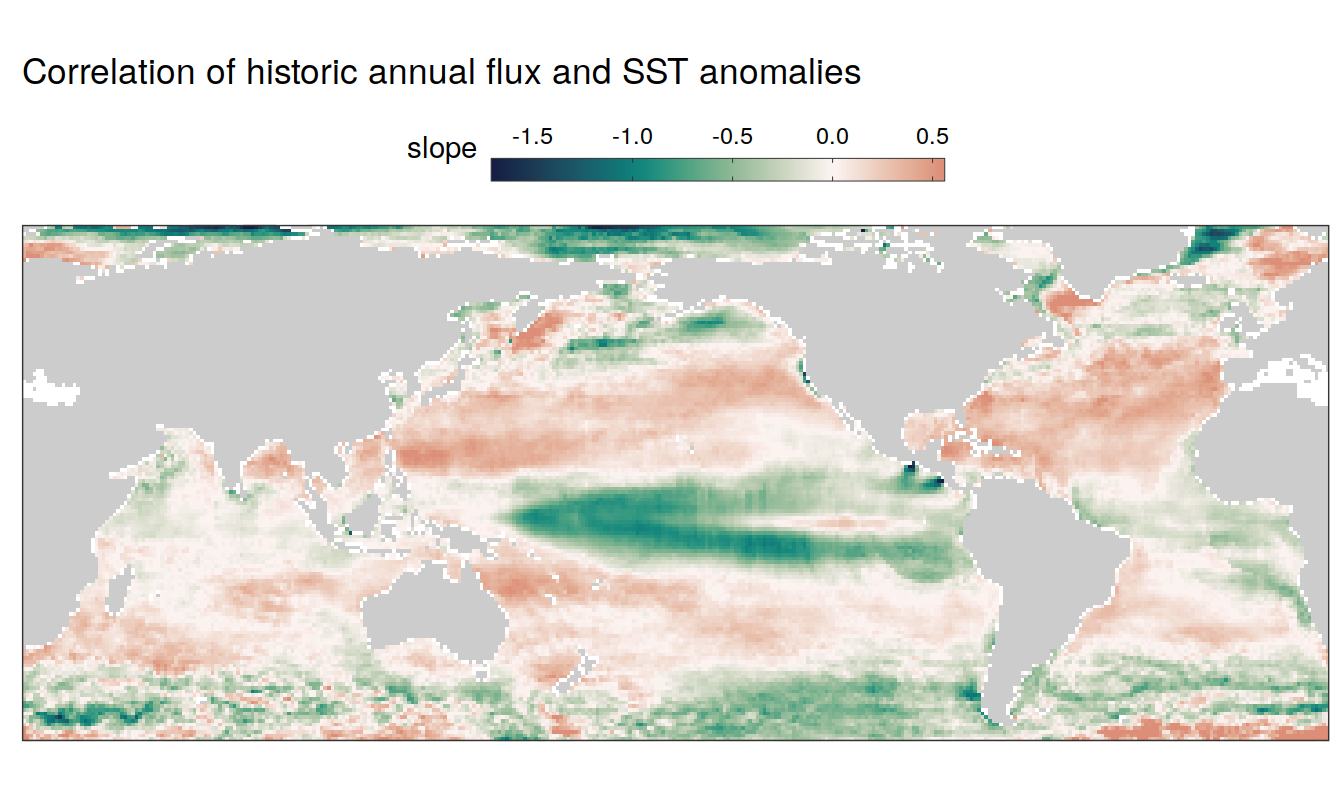
pco2_product_map_annual_anomaly_temperature_predict <-
inner_join(
pco2_product_map_annual_slope,
pco2_product_map_annual_anomaly %>%
filter(name %in% c("temperature", "fgco2")) %>%
select(lon, lat, name, resid, year) %>%
drop_na() %>%
arrange(desc(name)) %>%
pivot_wider(values_from = "resid")
)
pco2_product_map_annual_anomaly_temperature_predict <-
pco2_product_map_annual_anomaly_temperature_predict %>%
mutate(fgco2_predict = slope * temperature)
pco2_product_map_annual_anomaly_temperature_predict %>%
filter(year == 2023) %>%
select(-slope) %>%
pivot_longer(c(fgco2, temperature, fgco2_predict),
values_to = "resid") %>%
drop_na() %>%
# filter(name == "fgco2_predict") %>%
group_split(name) %>%
# head(1) %>%
map( ~ map +
geom_tile(data = .x,
aes(lon, lat, fill = resid)) +
labs(title = paste(2023,"anomaly")) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(),
legend.position = "top")
)[[1]]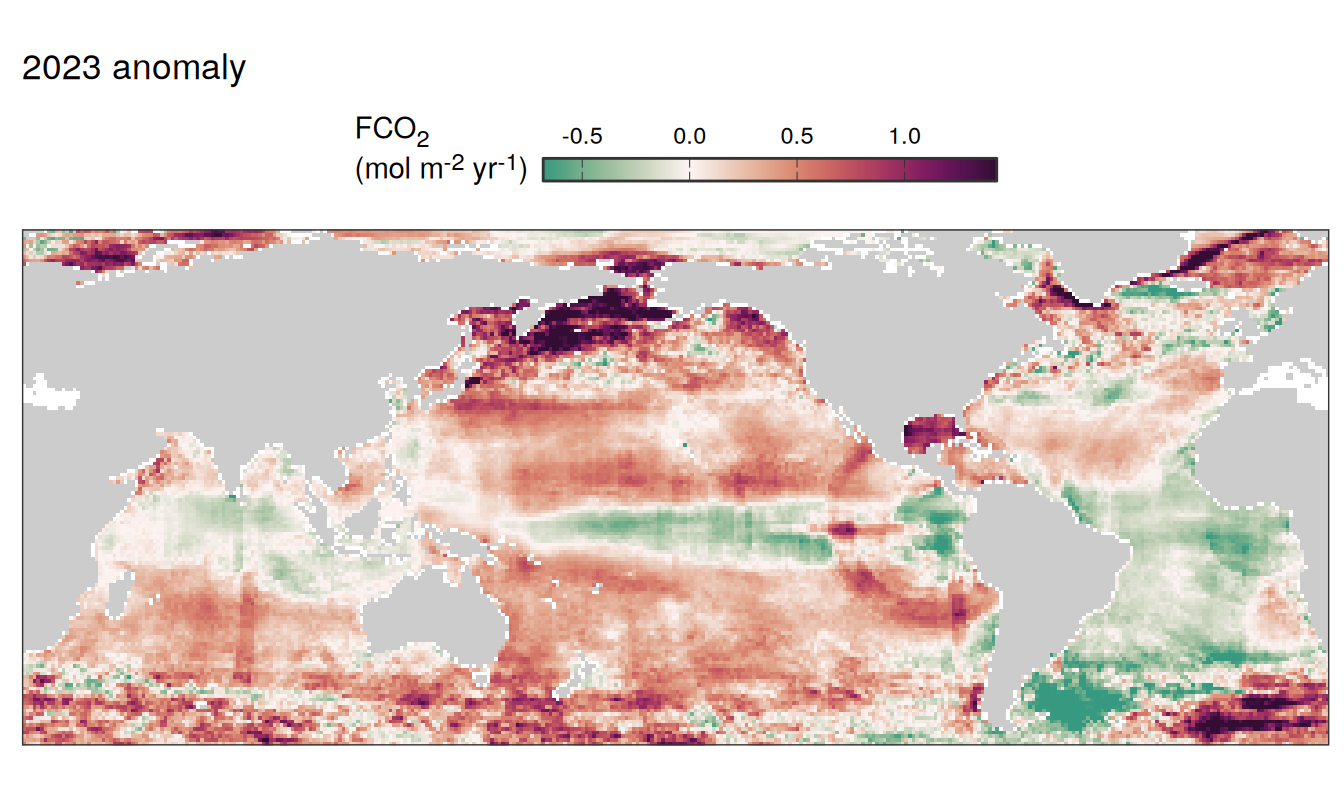
[[2]]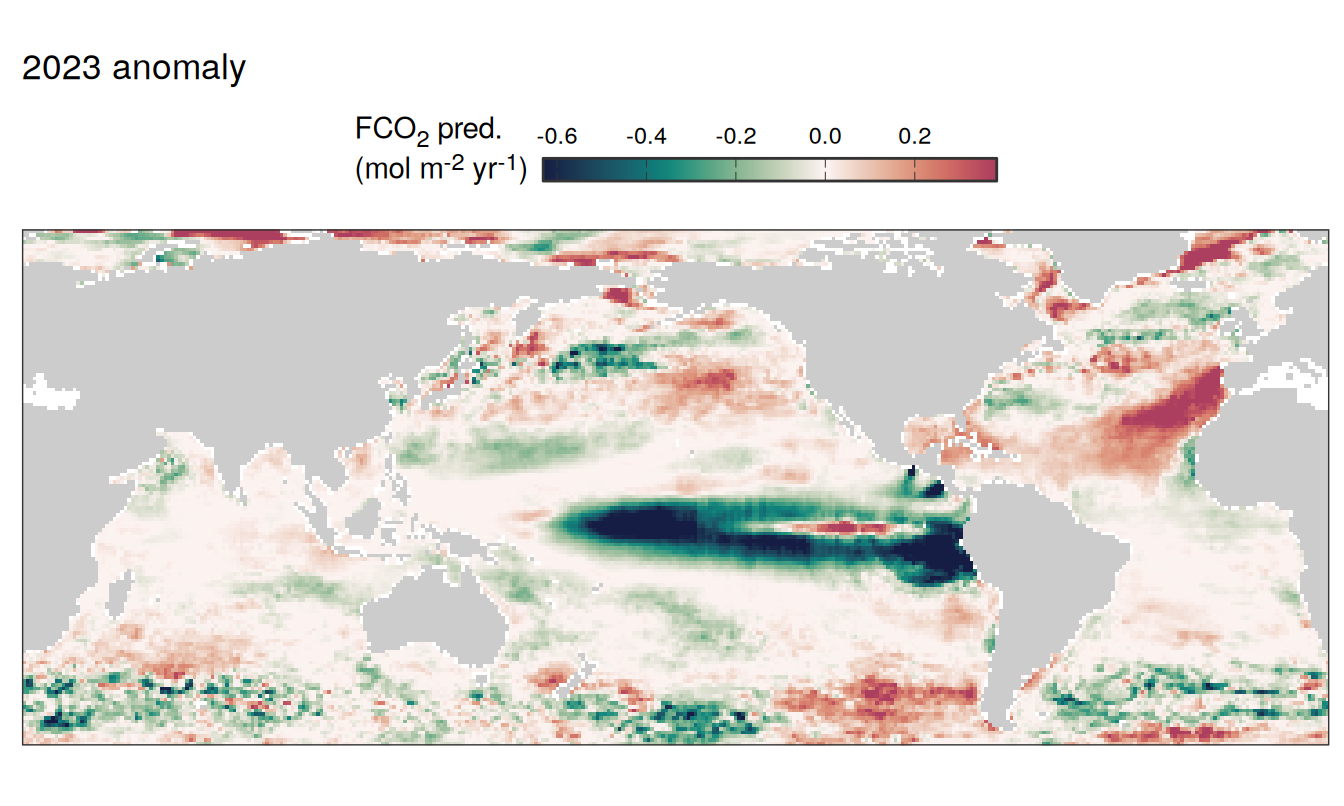
[[3]]
pco2_product_biome_annual_anomaly_temperature_predict <-
full_join(
pco2_product_map_annual_anomaly_temperature_predict %>%
select(lon, lat, year, temperature, fgco2, fgco2_predict),
biome_mask %>%
mutate(area = earth_surf(lat, lon))
)
pco2_product_biome_annual_anomaly_temperature_predict_global <-
pco2_product_biome_annual_anomaly_temperature_predict %>%
drop_na() %>%
mutate(fgco2_int = fgco2,
fgco2_predict_int = fgco2_predict) %>%
mutate(biome = case_when(str_detect(biome, "SO-SPSS|SO-ICE|Arctic") ~ "Polar",
TRUE ~ "Global non-polar")) %>%
filter(biome == "Global non-polar") %>%
select(-c(lon, lat)) %>%
group_by(year, biome) %>%
summarise(across(-c(fgco2_int, fgco2_predict_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(c(fgco2_int, fgco2_predict_int),
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_annual_anomaly_temperature_predict <-
pco2_product_biome_annual_anomaly_temperature_predict %>%
drop_na() %>%
mutate(fgco2_int = fgco2,
fgco2_predict_int = fgco2_predict) %>%
select(-c(lon, lat)) %>%
group_by(year, biome) %>%
summarise(across(-c(fgco2_int, fgco2_predict_int, area),
~ weighted.mean(., area, na.rm = TRUE)),
across(c(fgco2_int, fgco2_predict_int),
~ sum(. * area, na.rm = TRUE) * 12.01 * 1e-15)) %>%
ungroup()
pco2_product_biome_annual_anomaly_temperature_predict <-
bind_rows(pco2_product_biome_annual_anomaly_temperature_predict_global,
pco2_product_biome_annual_anomaly_temperature_predict)
rm(pco2_product_biome_annual_anomaly_temperature_predict_global)
pco2_product_biome_annual_anomaly_temperature_predict <-
pco2_product_biome_annual_anomaly_temperature_predict %>%
filter(!is.na(biome))
pco2_product_biome_annual_anomaly_temperature_predict %>%
select(year, biome, fgco2_int, fgco2_predict_int) %>%
filter(biome == "Global non-polar") %>%
pivot_longer(starts_with("fgco2"),
values_to = "resid") %>%
ggplot(aes(year, resid, col = name)) +
geom_path() +
geom_point()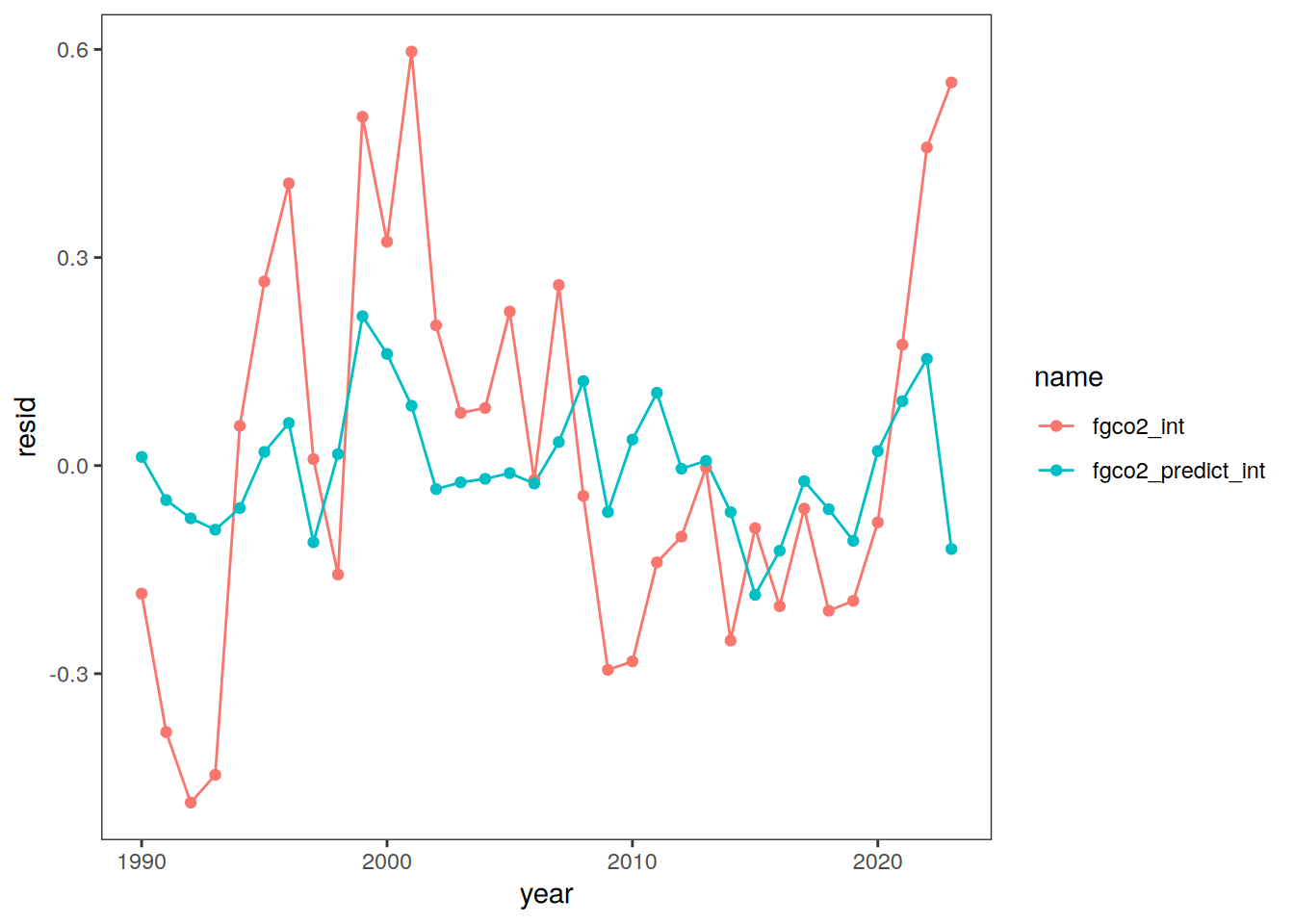
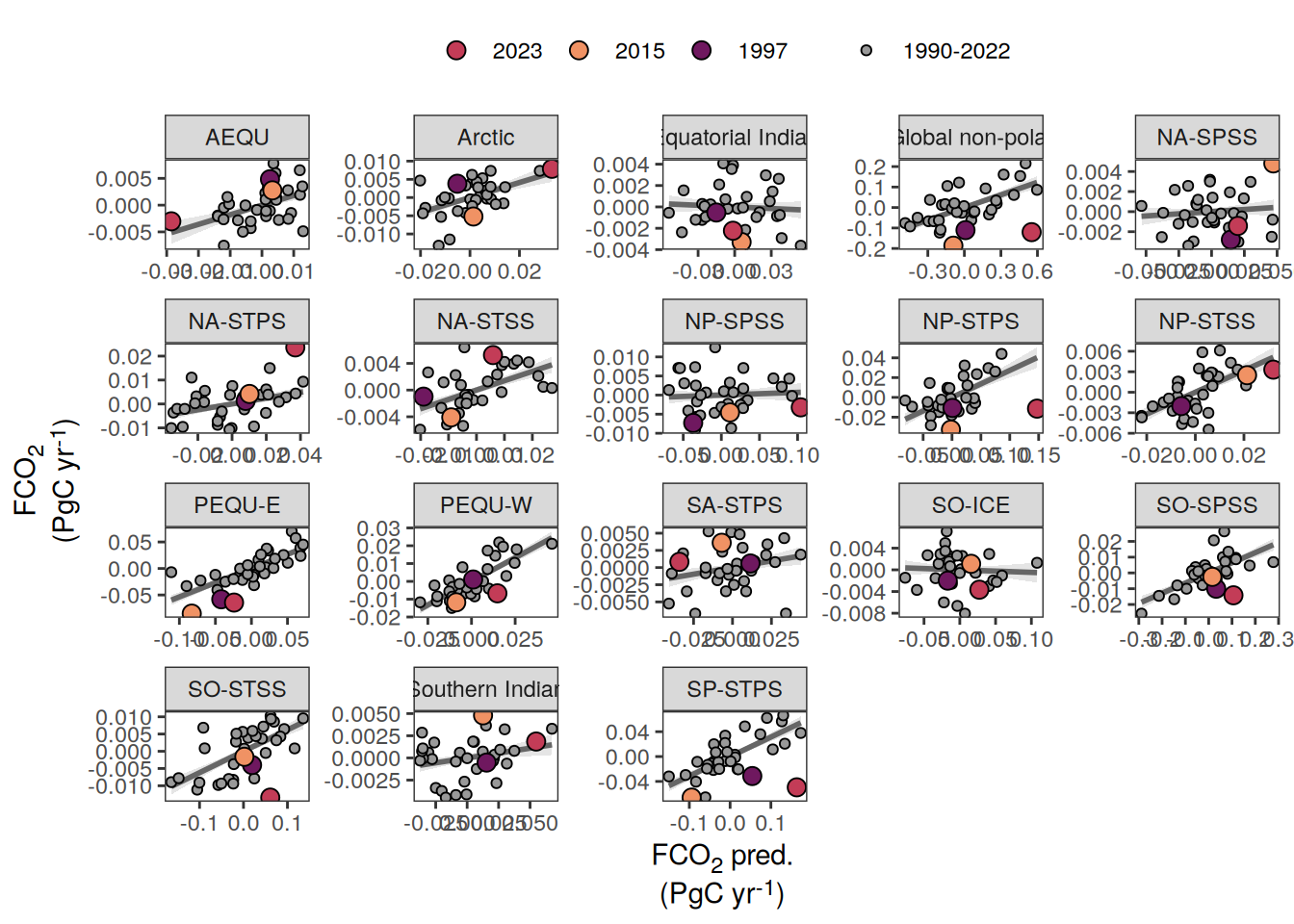
pco2_product_biome_annual_anomaly_temperature_predict %>%
ggplot(aes(fgco2_int, fgco2_predict_int)) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_point(data = . %>% filter(!year %in% c(2023, 1997, 2015)),
aes(fill = "1990-2022"),
shape = 21) +
scale_color_manual(values = "grey60", name = "X") +
scale_fill_manual(values = "grey60", name = "X") +
new_scale_fill() +
new_scale_color() +
geom_point(
data = . %>% filter(year %in% c(2023, 1997, 2015)),
aes(fill = as.factor(year)),
shape = 21,
size = 3
) +
scale_fill_manual(
values = rev(warm_cool_gradient[c(17,13,20)]),
guide = guide_legend(reverse = TRUE,
order = 2)
) +
scale_color_manual(
values = rev(warm_cool_gradient[c(17,13,20)]),
guide = guide_legend(reverse = TRUE,
order = 2)
) +
labs(y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks("fgco2_predict_int")$i_legend_title) +
facet_wrap(~ biome, scales = "free") +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)
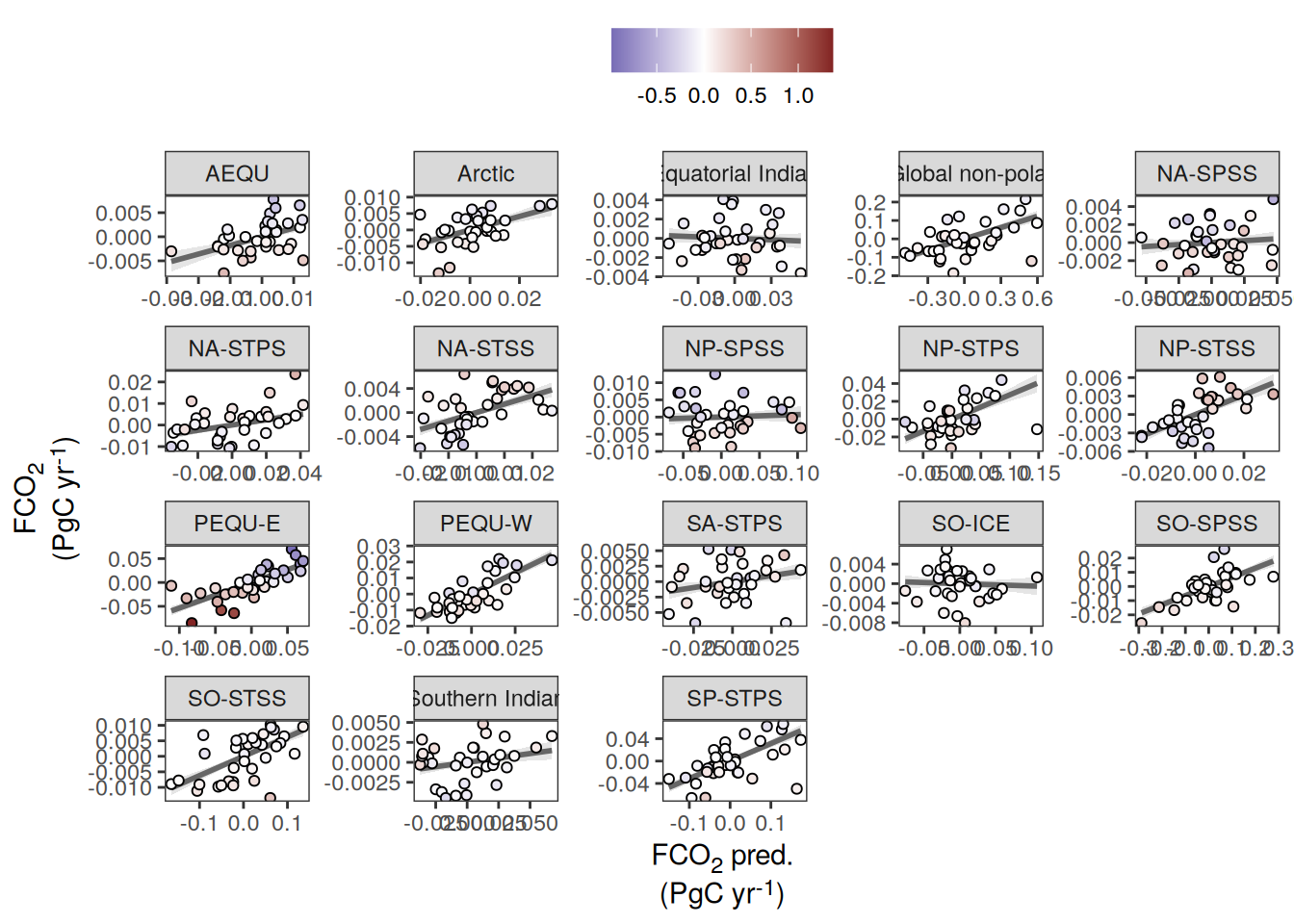
pco2_product_biome_annual_anomaly_temperature_predict %>%
ggplot(aes(fgco2_int, fgco2_predict_int)) +
geom_smooth(
data = . %>% filter(year != 2023),
method = "lm",
fill = "grey",
col = "grey40",
fullrange = TRUE,
level = 0.68
) +
geom_point(aes(fill = temperature),
shape = 21) +
scale_fill_divergent() +
labs(y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks("fgco2_predict_int")$i_legend_title) +
facet_wrap(~ biome, scales = "free") +
theme(
axis.title.x = element_markdown(),
axis.title.y = element_markdown(),
legend.title = element_blank(),
legend.position = "top"
)
pco2_product_map_annual_anomaly %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_map_annual_anomaly.csv"
)
)
pco2_product_map_annual_anomaly_temperature_predict %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_map_annual_anomaly_temperature_predict.csv"
)
)
pco2_product_biome_annual_anomaly_temperature_predict %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_annual_anomaly_temperature_predict.csv"
)
)
rm(pco2_product_map_annual_anomaly,
pco2_product_map_annual_slope,
pco2_product_map_annual_anomaly_temperature_predict,
pco2_product_biome_annual_anomaly_temperature_predict)
gc()Monthly means
2023 absolute
pco2_product_map_monthly_anomaly <-
pco2_product_map_monthly %>%
drop_na() %>%
anomaly_determination(lon, lat, month)
pco2_product_map_monthly_anomaly <-
pco2_product_map_monthly_anomaly %>%
drop_na()
pco2_product_map_monthly_anomaly %>%
filter(year == 2023, !(name %in% name_divergent)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = value)) +
labs(title = paste("Monthly means", 2023)) +
scale_fill_viridis_c(name = labels_breaks(.x %>% distinct(name))) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap(~ month, ncol = 2)
)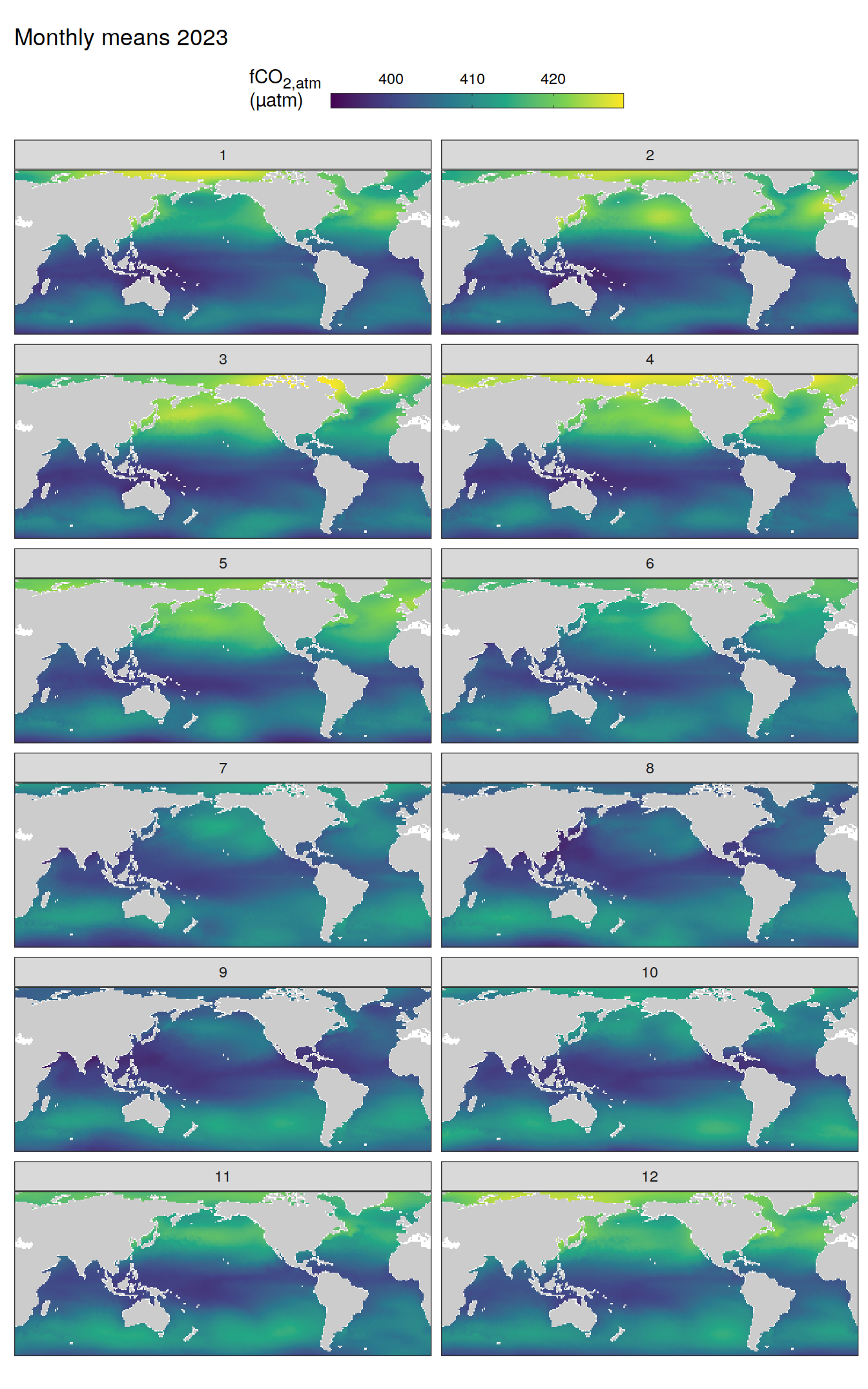
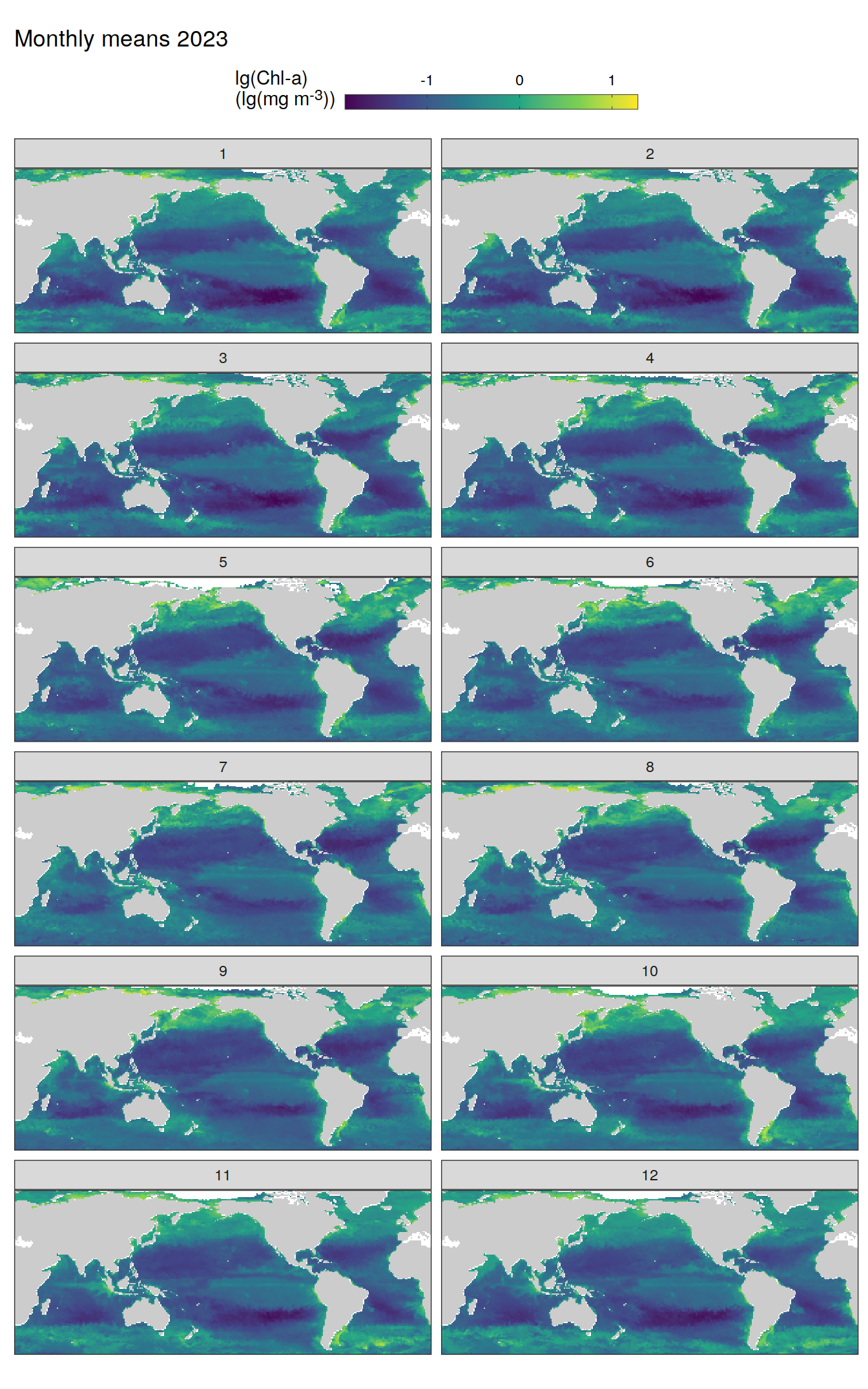
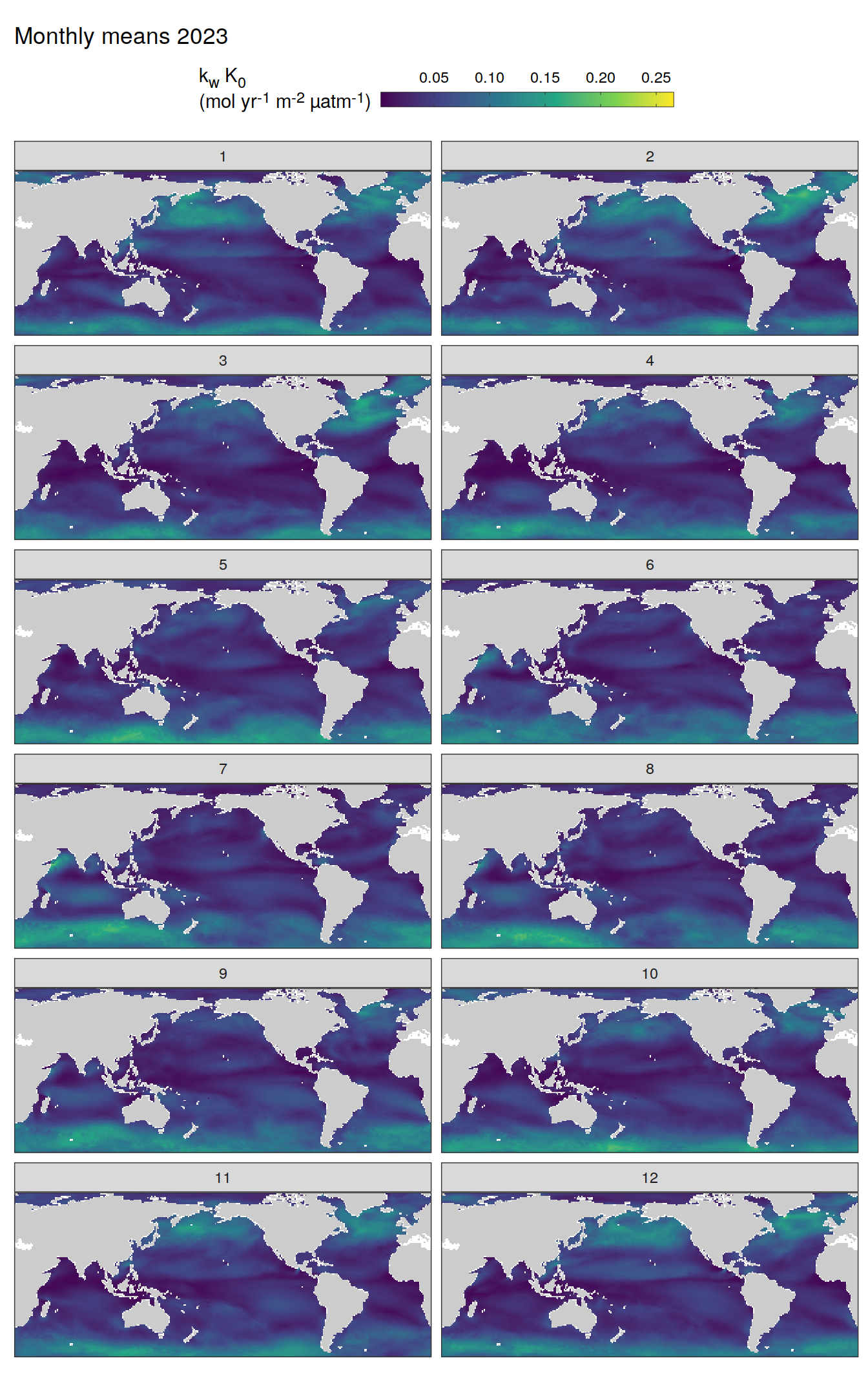
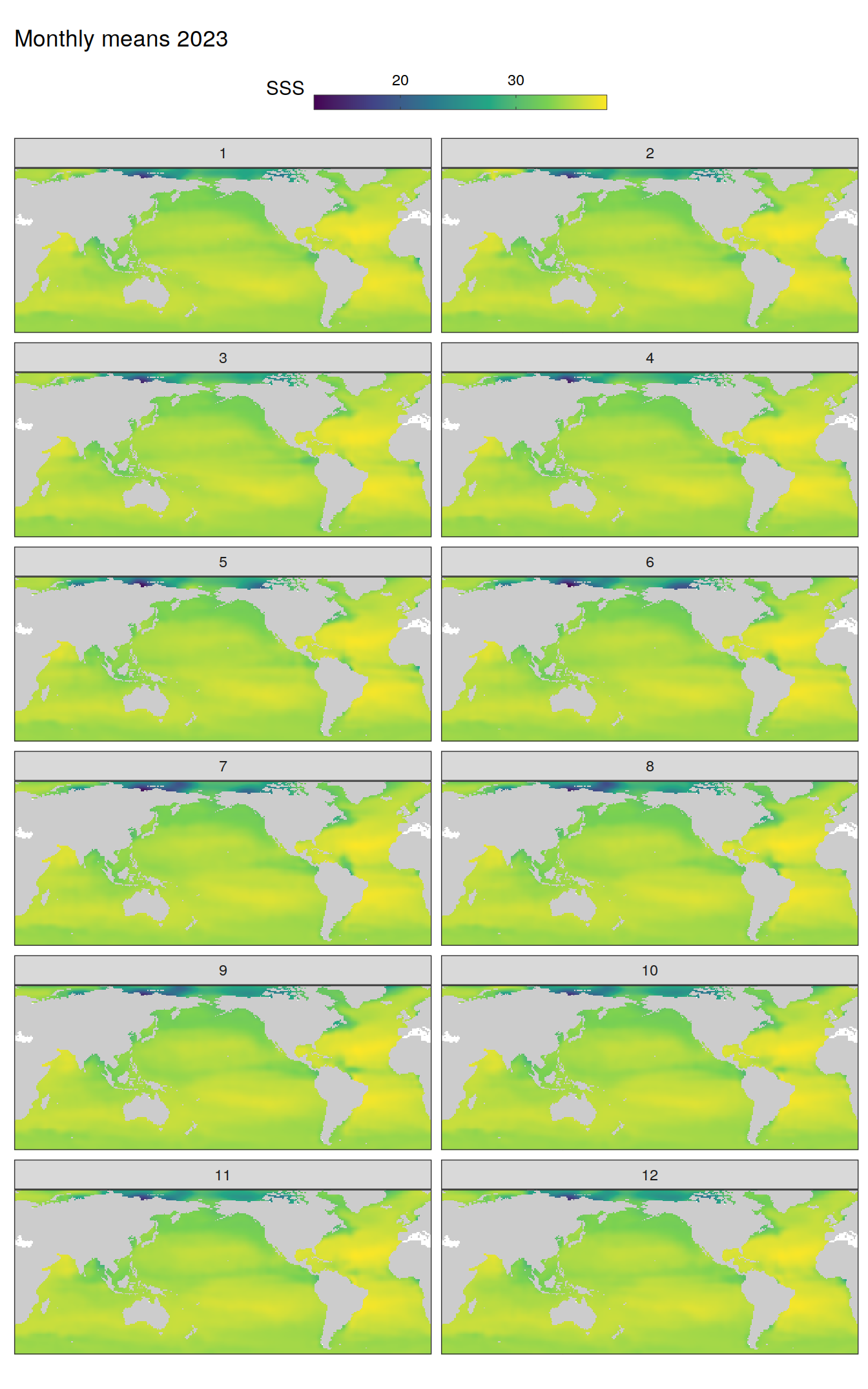
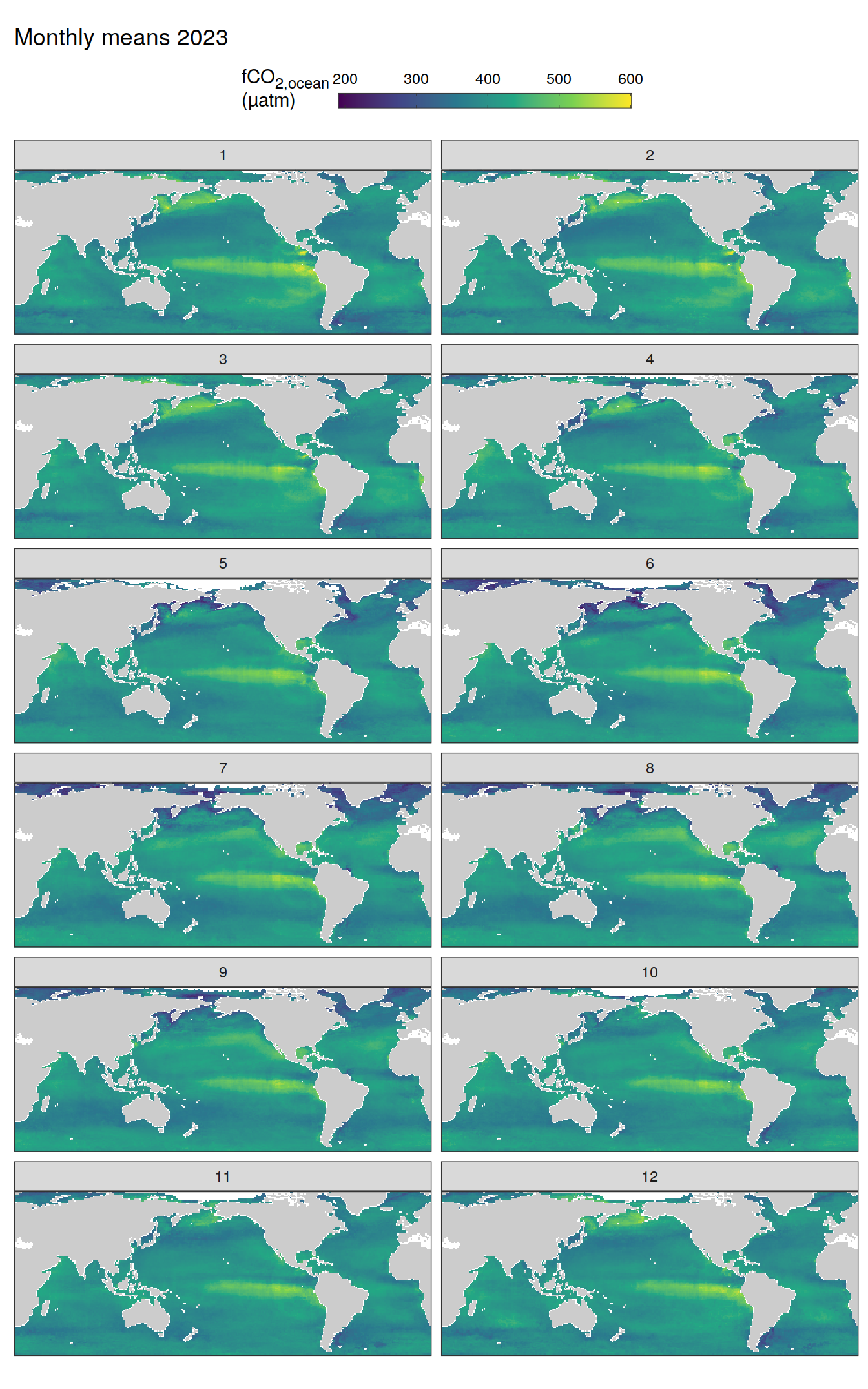
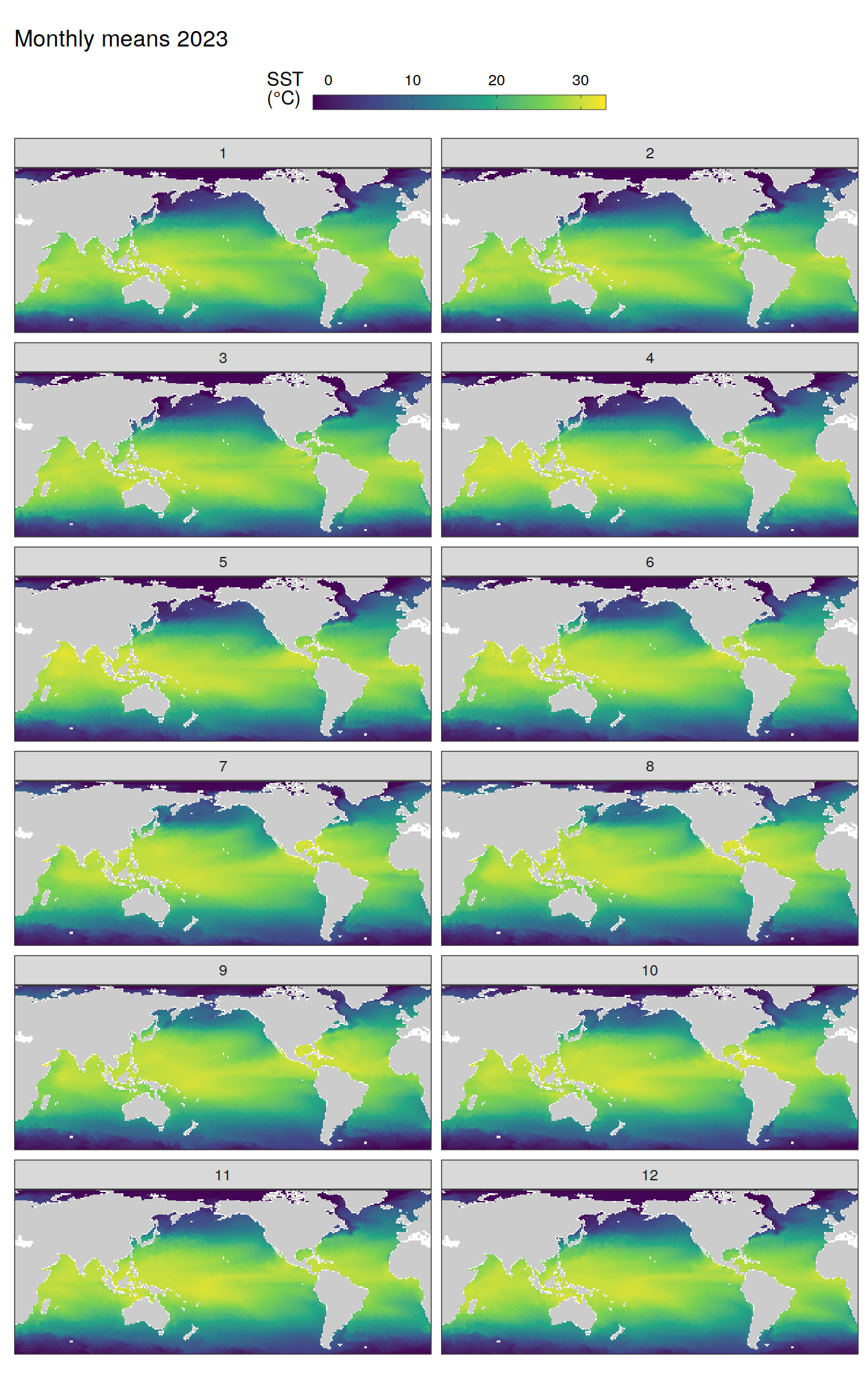
pco2_product_map_monthly_anomaly %>%
filter(year == 2023, name %in% name_divergent) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = value)) +
labs(title = paste("Monthly means", 2023)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$value, .01), quantile(.x$value, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap( ~ month, ncol = 2)
)

Trends
pco2_product_map_monthly_anomaly %>%
group_by(name) %>%
filter(year %in% c(min(year), max(year))) %>%
ungroup() %>%
select(-c(value, resid)) %>%
arrange(year) %>%
group_by(lon, lat, name, month) %>%
mutate(change = fit - lag(fit),
period = paste(lag(year), year, sep = "-")) %>%
ungroup() %>%
filter(!is.na(change)) %>%
group_split(name) %>%
head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = change)) +
labs(title = paste("Change: ", .x$period)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$change, .01), quantile(.x$change, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap(~ month, ncol = 2)
)2023 anomaly
pco2_product_map_monthly_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(1) %>%
map(
~ map +
geom_tile(data = .x, aes(lon, lat, fill = resid)) +
labs(title = paste(2023, "anomaly")) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top") +
facet_wrap( ~ month, ncol = 2)
)
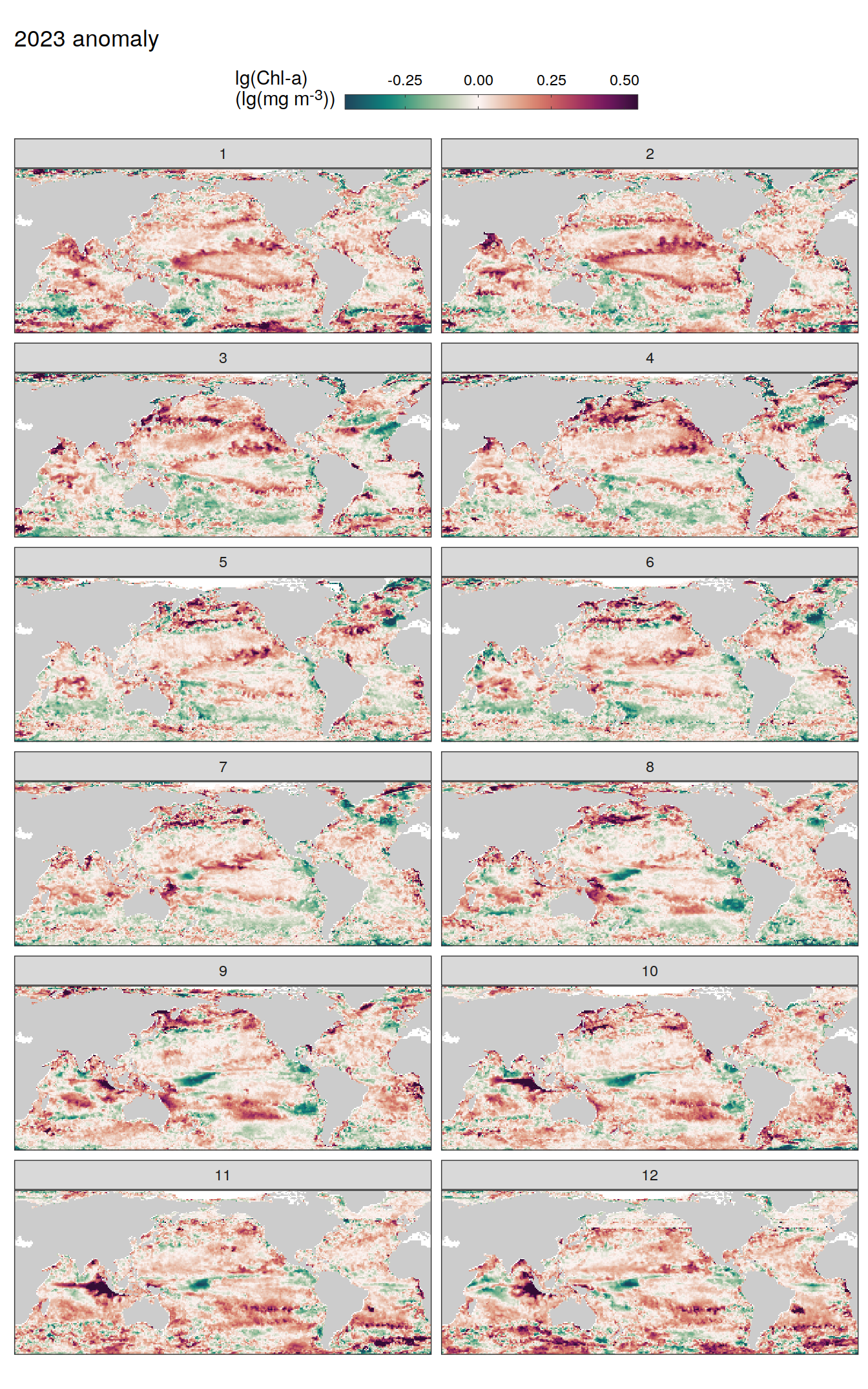
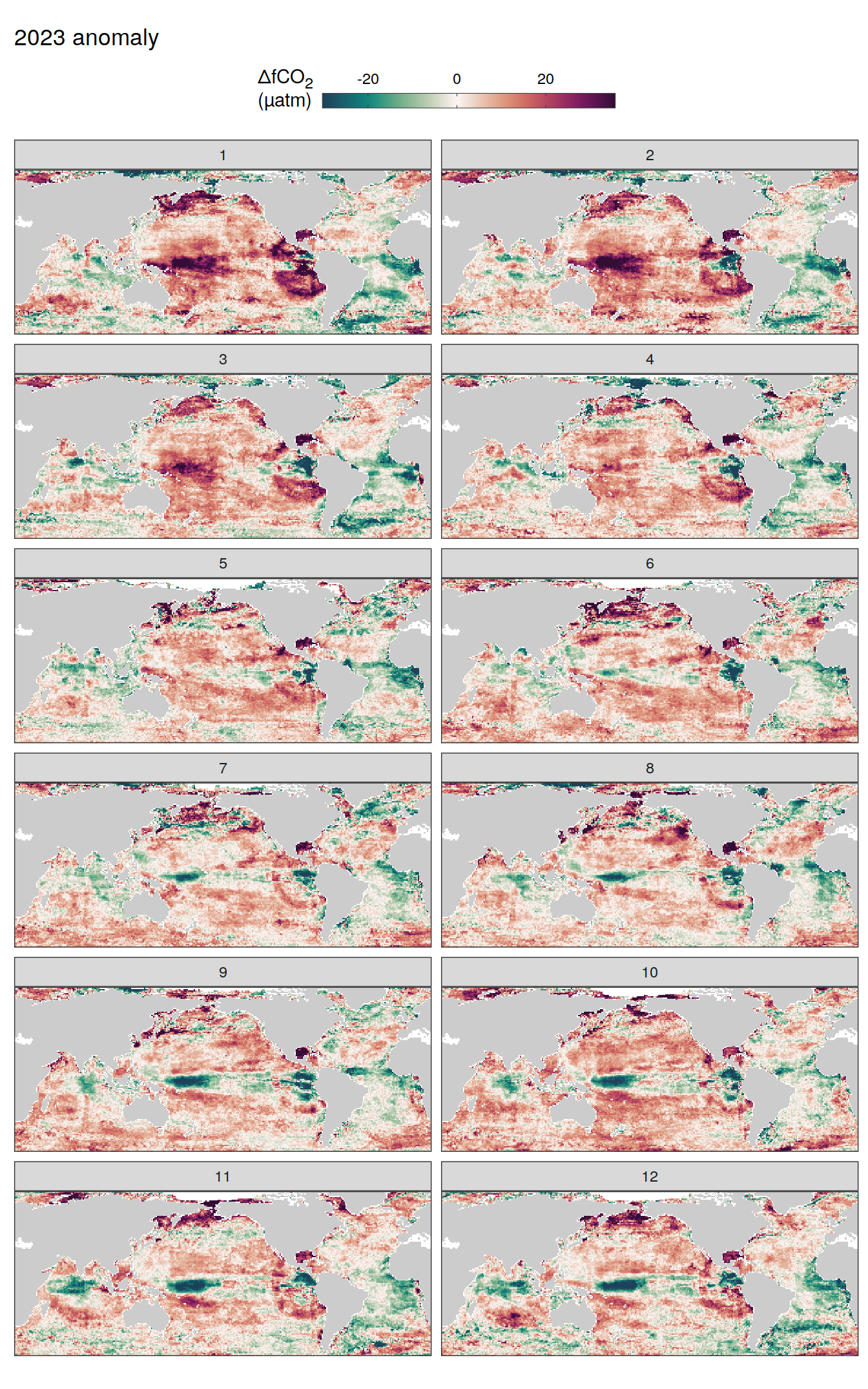
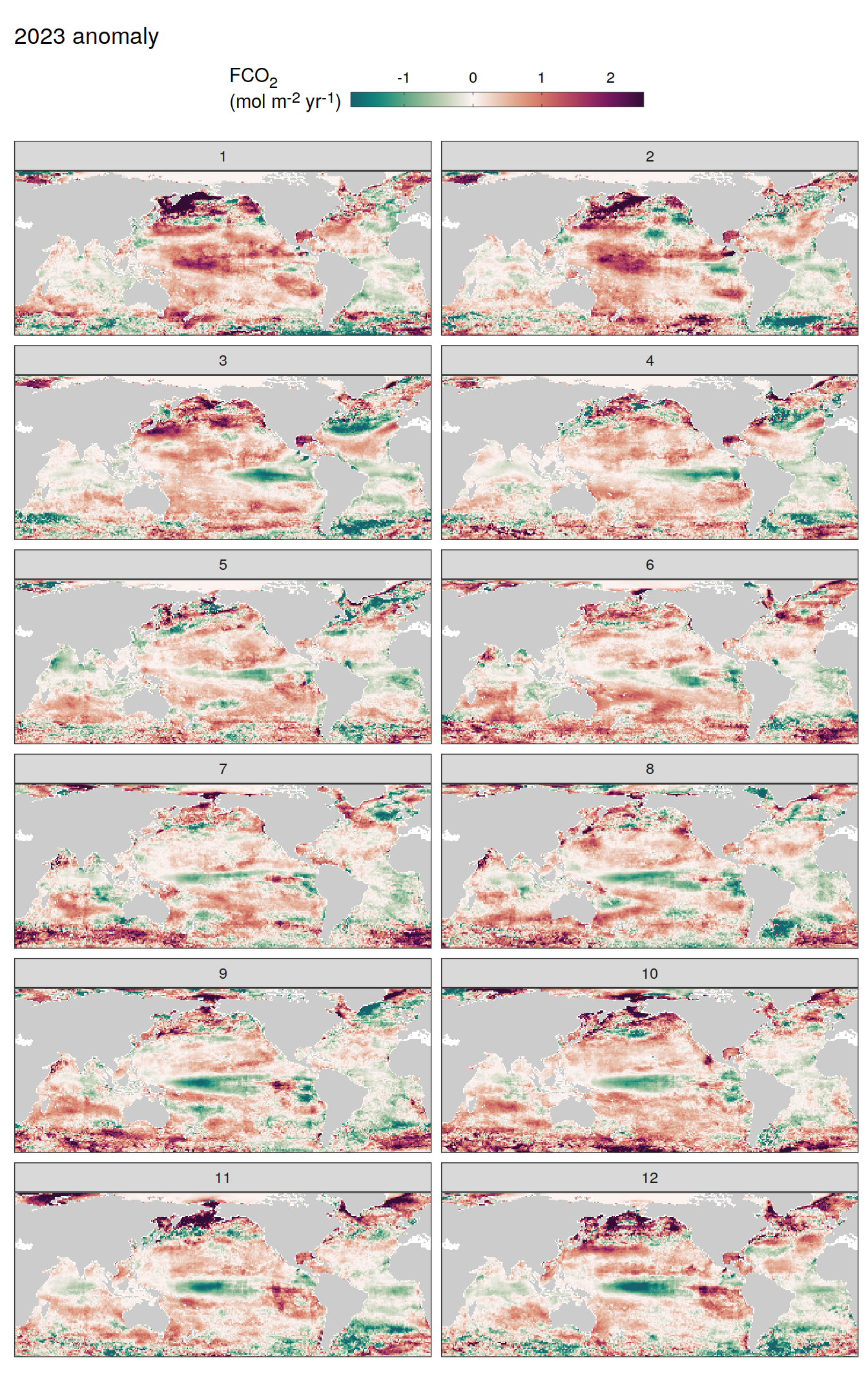
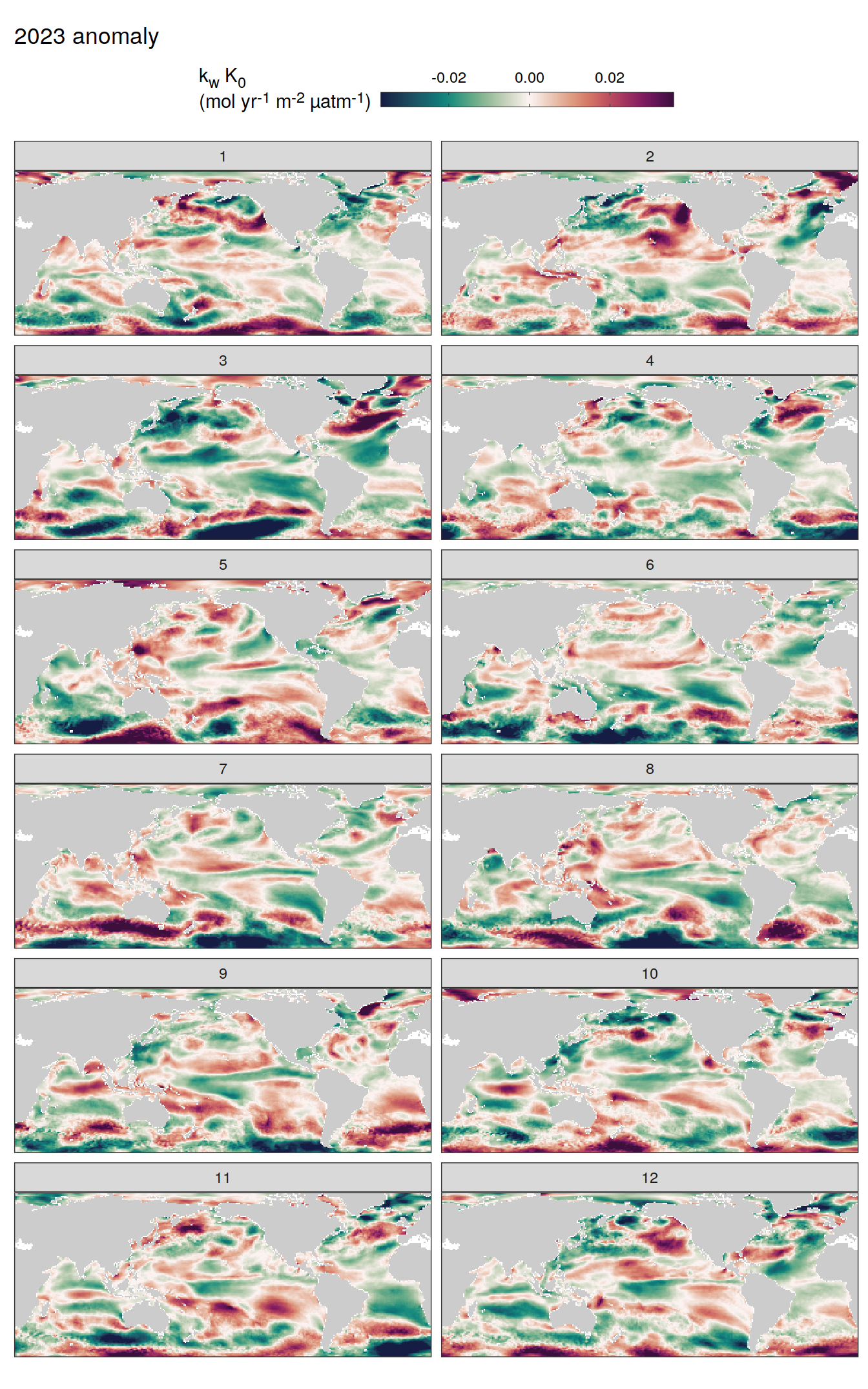
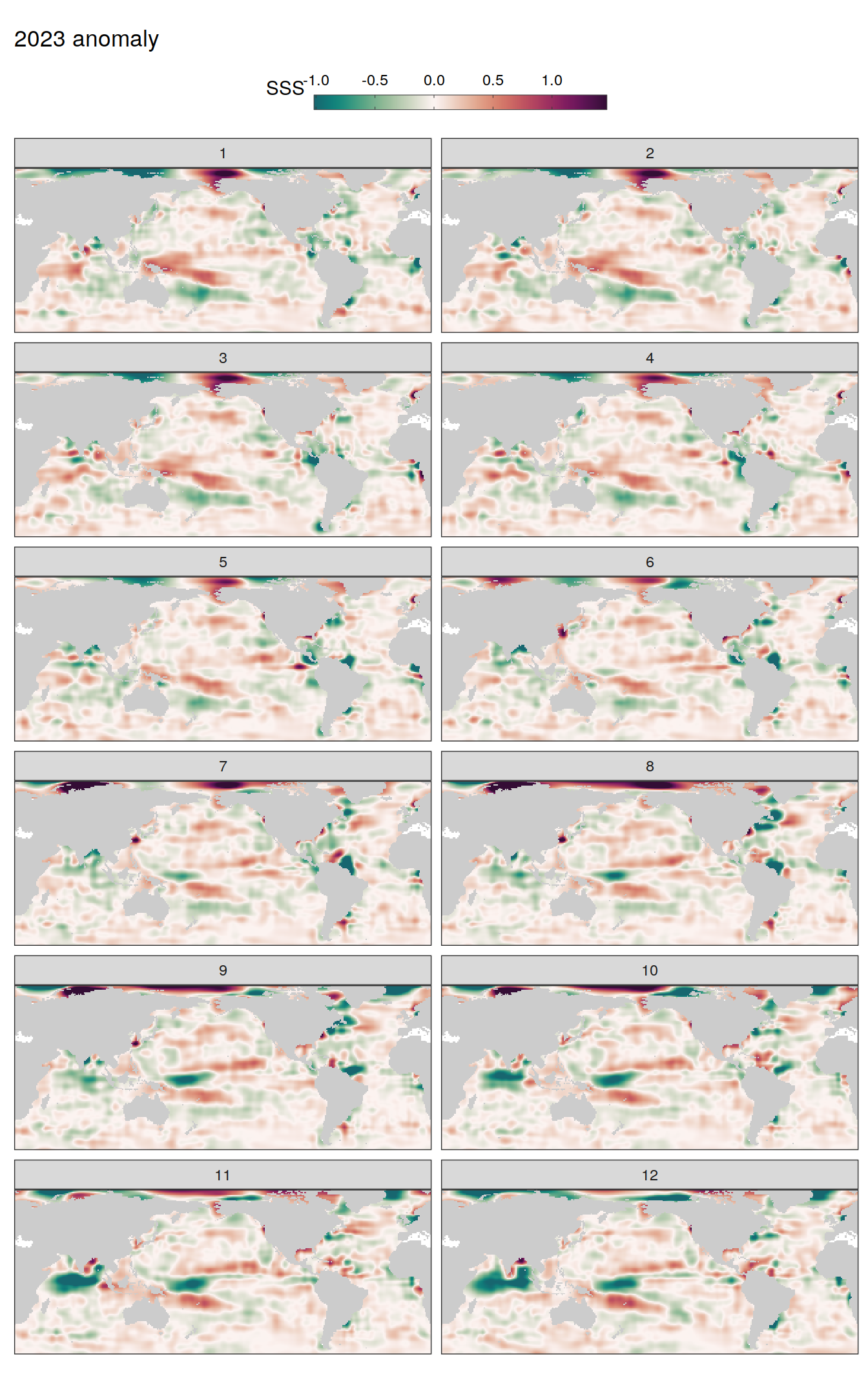
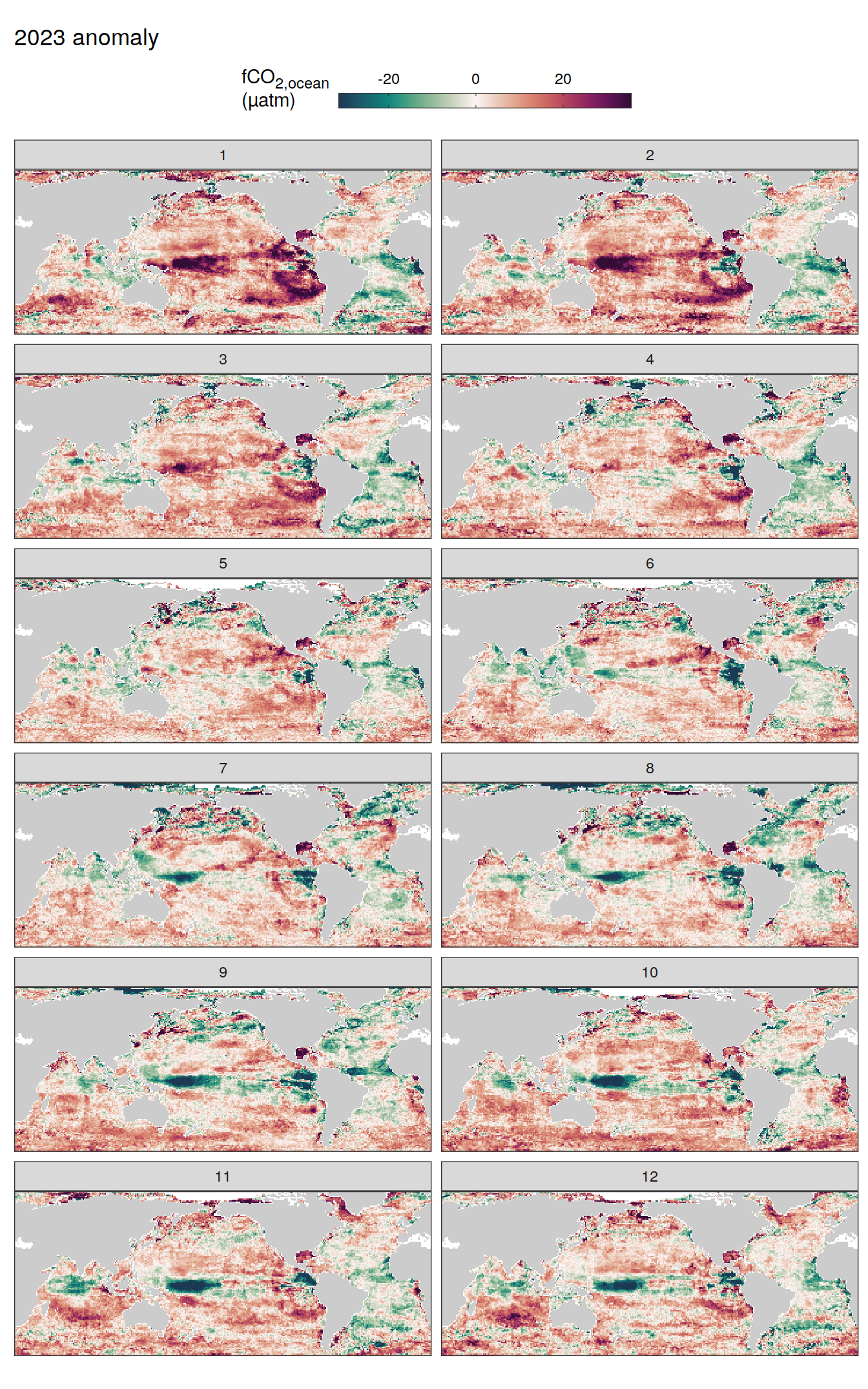
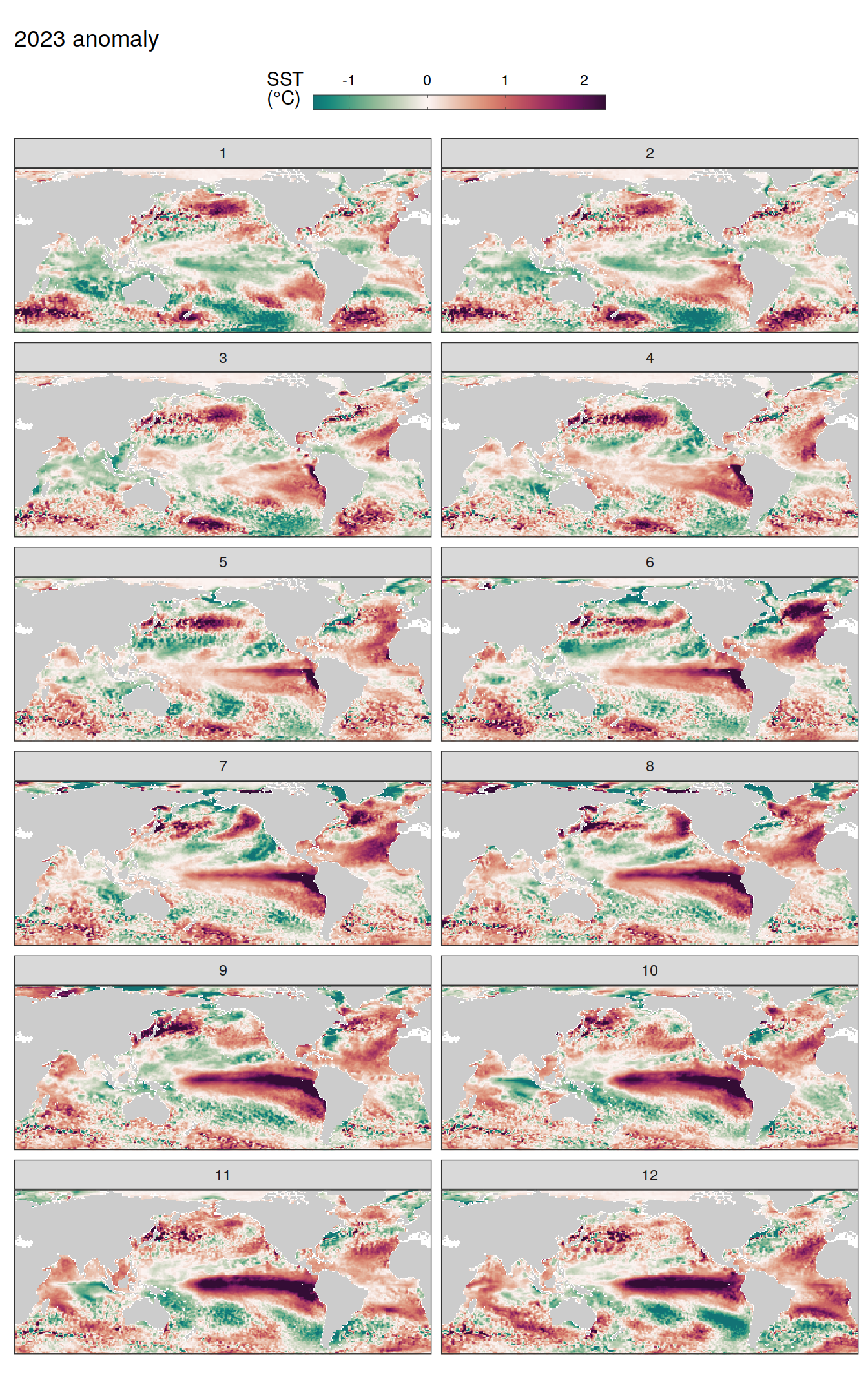
fCO2 decomposition
pco2_product_map_monthly_fCO2_decomposition <-
fco2_decomposition(pco2_product_map_monthly_anomaly,
year, month, lon, lat)
# pco2_product_map_monthly_fCO2_decomposition %>%
# filter(year == 2023) %>%
# mutate(product == "pco2 product") %>%
# group_split(product) %>%
# head(1) %>%
# map(
# ~ map +
# geom_tile(data = .x,
# aes(lon, lat, fill = resid)) +
# labs(title = .x$product) +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks("sfco2"),
# limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
# oob = squish
# ) +
# facet_grid(month ~ name,
# labeller = labeller(name = x_axis_labels)) +
# guides(
# fill = guide_colorbar(
# barheight = unit(0.3, "cm"),
# barwidth = unit(6, "cm"),
# ticks = TRUE,
# ticks.colour = "grey20",
# frame.colour = "grey20",
# label.position = "top",
# direction = "horizontal"
# )
# ) +
# theme(legend.title = element_markdown(),
# legend.position = "top")
# )pco2_product_map_annual_fCO2_decomposition <-
pco2_product_map_monthly_fCO2_decomposition %>%
select(year, lat, lon, name, resid) %>%
fgroup_by(year, lat, lon, name) %>%
fmean()
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4288211 229.1 52704091 2814.8 160840118 8589.8
Vcells 2225855720 16982.0 5170874616 39450.7 5170874616 39450.7map +
geom_tile(data = pco2_product_map_annual_fCO2_decomposition %>%
filter(year == 2023), aes(lon, lat, fill = resid)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("sfco2"),
limits = c(
quantile(pco2_product_map_annual_fCO2_decomposition$resid, .01),
quantile(pco2_product_map_annual_fCO2_decomposition$resid, .99)
),
oob = squish
) +
facet_wrap(~ name,
ncol = 2,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")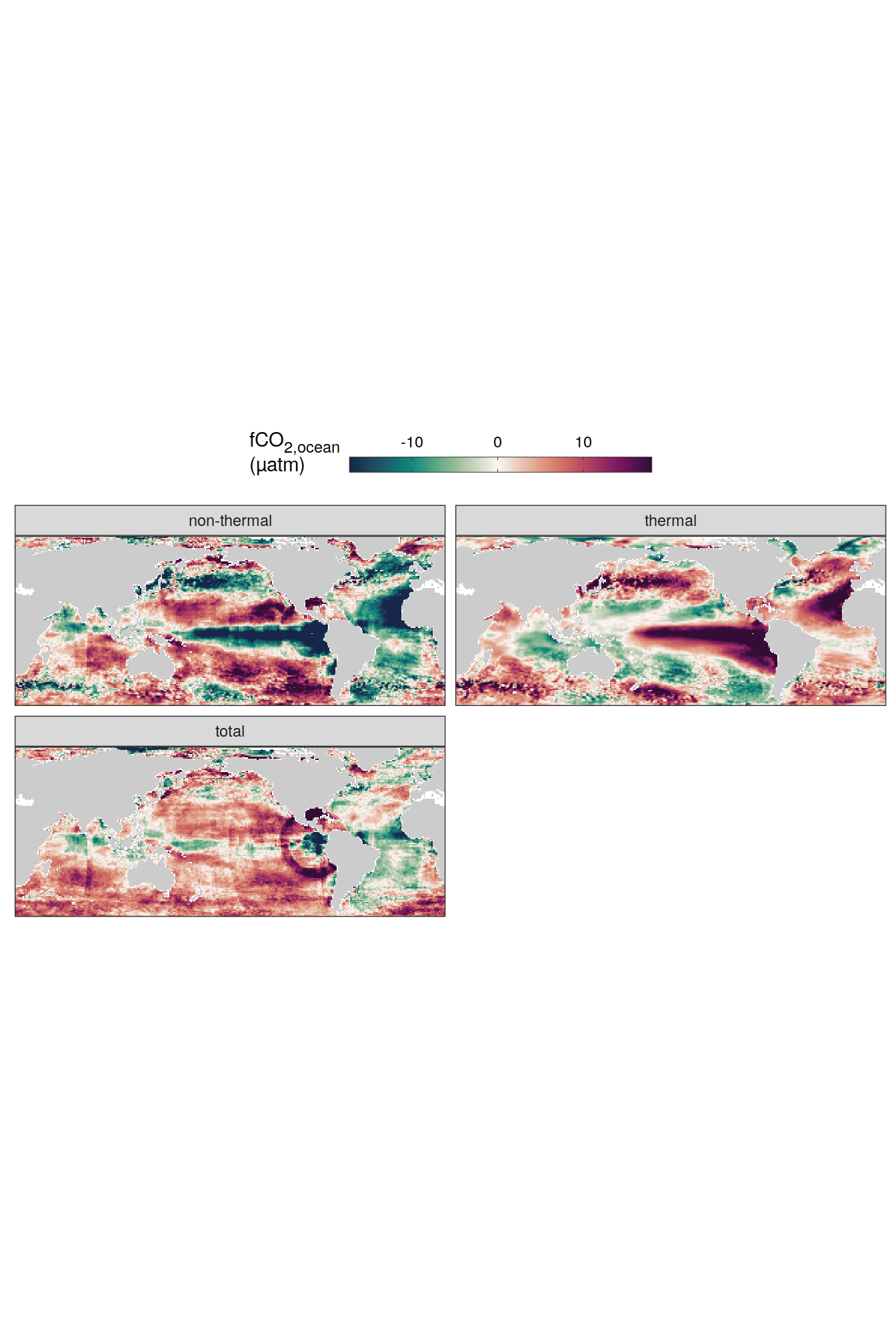
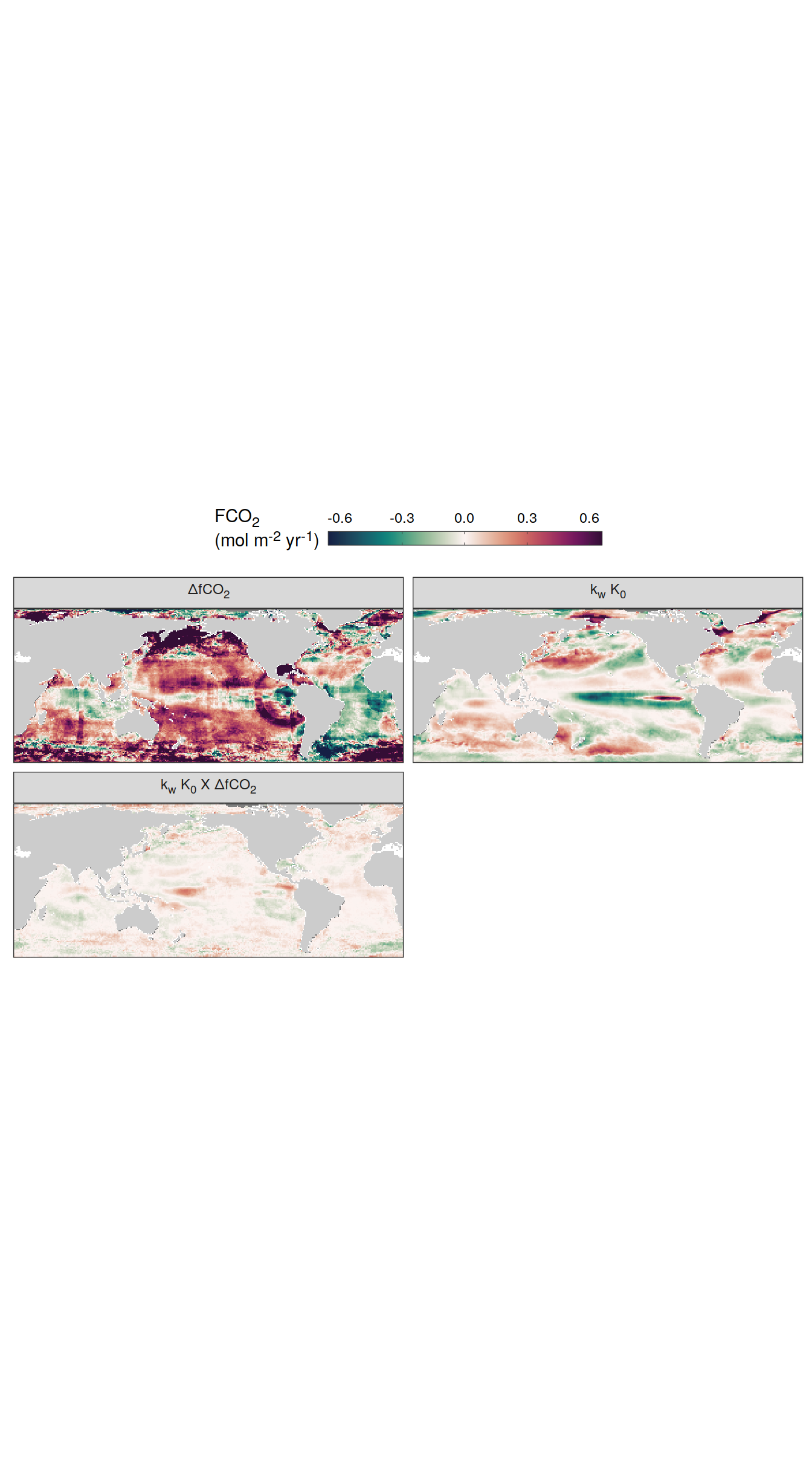
Flux attribution
pco2_product_map_monthly_flux_attribution <-
flux_attribution(pco2_product_map_monthly_anomaly,
year, month, lon, lat)
# pco2_product_map_monthly_flux_attribution %>%
# filter(year == 2023) %>%
# drop_na() %>%
# mutate(product == "pco2 product") %>%
# group_split(product) %>%
# head(1) %>%
# map(
# ~ map +
# geom_tile(data = .x,
# aes(lon, lat, fill = resid)) +
# labs(subtitle = .x$product) +
# scale_fill_gradientn(
# colours = cmocean("curl")(100),
# rescaler = ~ scales::rescale_mid(.x, mid = 0),
# name = labels_breaks("fgco2"),
# limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
# oob = squish
# ) +
# theme(legend.title = element_markdown(),
# legend.position = "bottom") +
# facet_grid(month ~ name,
# labeller = labeller(name = x_axis_labels)) +
# guides(
# fill = guide_colorbar(
# barheight = unit(0.3, "cm"),
# barwidth = unit(6, "cm"),
# ticks = TRUE,
# ticks.colour = "grey20",
# frame.colour = "grey20",
# label.position = "top",
# direction = "horizontal"
# )
# ) +
# theme(legend.title = element_markdown(),
# legend.position = "top",
# strip.text.x.top = element_markdown())
# )pco2_product_map_annual_flux_attribution <-
pco2_product_map_monthly_flux_attribution %>%
group_by(year, lat, lon, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup()
map +
geom_tile(data = pco2_product_map_annual_flux_attribution %>%
filter(year == 2023), aes(lon, lat, fill = resid)) +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks("fgco2"),
limits = c(
quantile(pco2_product_map_annual_flux_attribution$resid, .01, na.rm = TRUE),
quantile(pco2_product_map_annual_flux_attribution$resid, .99, na.rm = TRUE)
),
oob = squish
) +
theme(legend.title = element_markdown(), legend.position = "bottom") +
facet_wrap(~ name,
ncol = 2,
labeller = labeller(name = x_axis_labels)) +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(
legend.title = element_markdown(),
legend.position = "top",
strip.text.x.top = element_markdown()
)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4306256 230.0 42163273 2251.8 160840118 8589.8
Vcells 2488151648 18983.1 5170874616 39450.7 5170874616 39450.7pco2_product_map_monthly_anomaly %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_map_monthly_anomaly.csv"
)
)
pco2_product_map_annual_flux_attribution %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_map_annual_flux_attribution.csv"
)
)
pco2_product_map_annual_fCO2_decomposition %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_map_annual_fCO2_decomposition.csv"
)
)
pco2_product_map_monthly_flux_attribution %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_map_monthly_flux_attribution.csv"
)
)
pco2_product_map_monthly_fCO2_decomposition %>%
filter(year == 2023) %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_map_monthly_fCO2_decomposition.csv"
)
)
rm(pco2_product_map_annual_flux_attribution,
pco2_product_map_annual_fCO2_decomposition)
gc()Hovmoeller plots
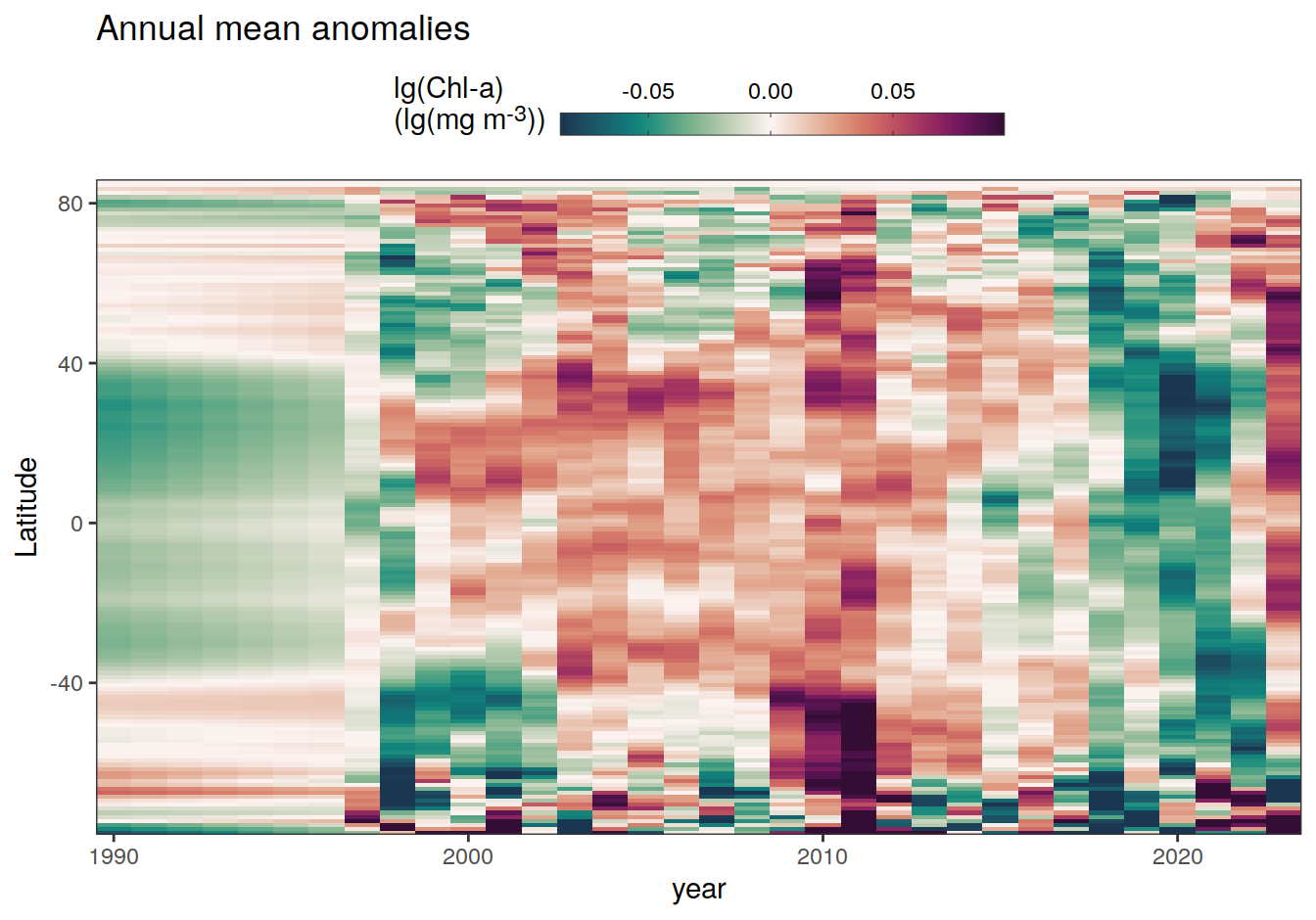
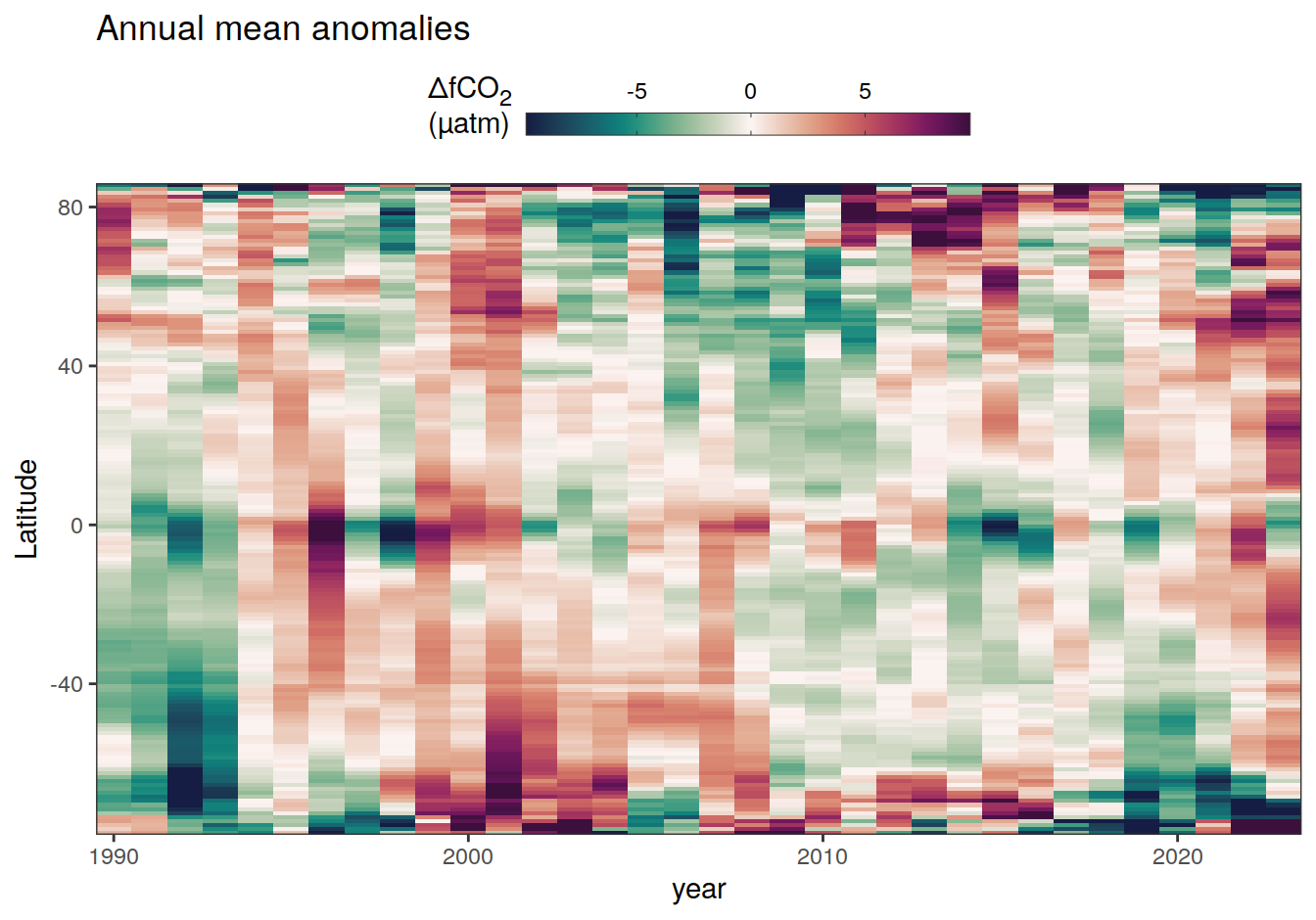
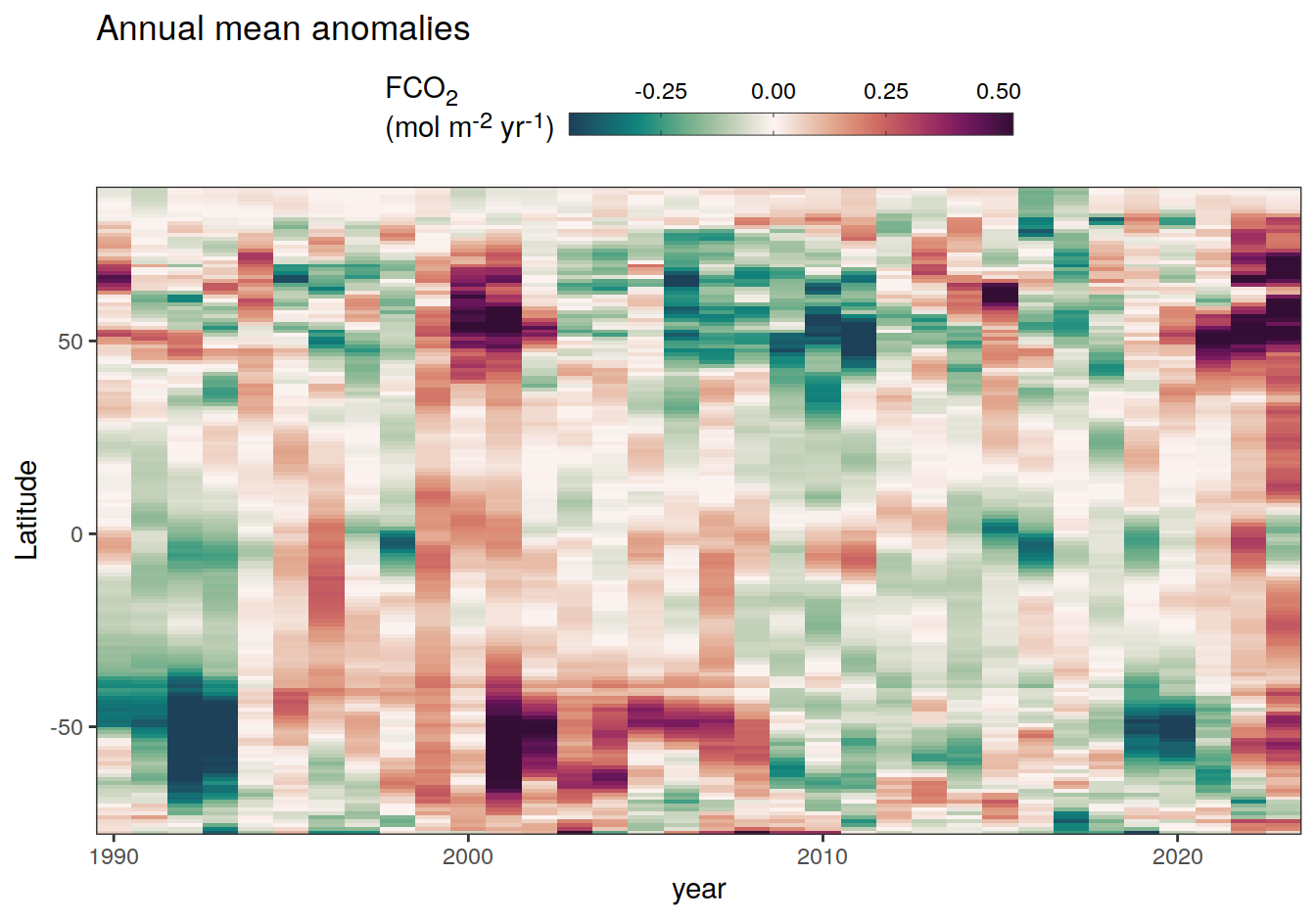
The following Hovmoeller plots show the anomalies from the prediction of the linear/quadratic fits.
Hovmoeller plots are first presented as annual means, and than as monthly means. Note that the predictions for the monthly Hovmoeller plots are done individually for each month, such the mean seasonal anomaly from the annual mean is removed.
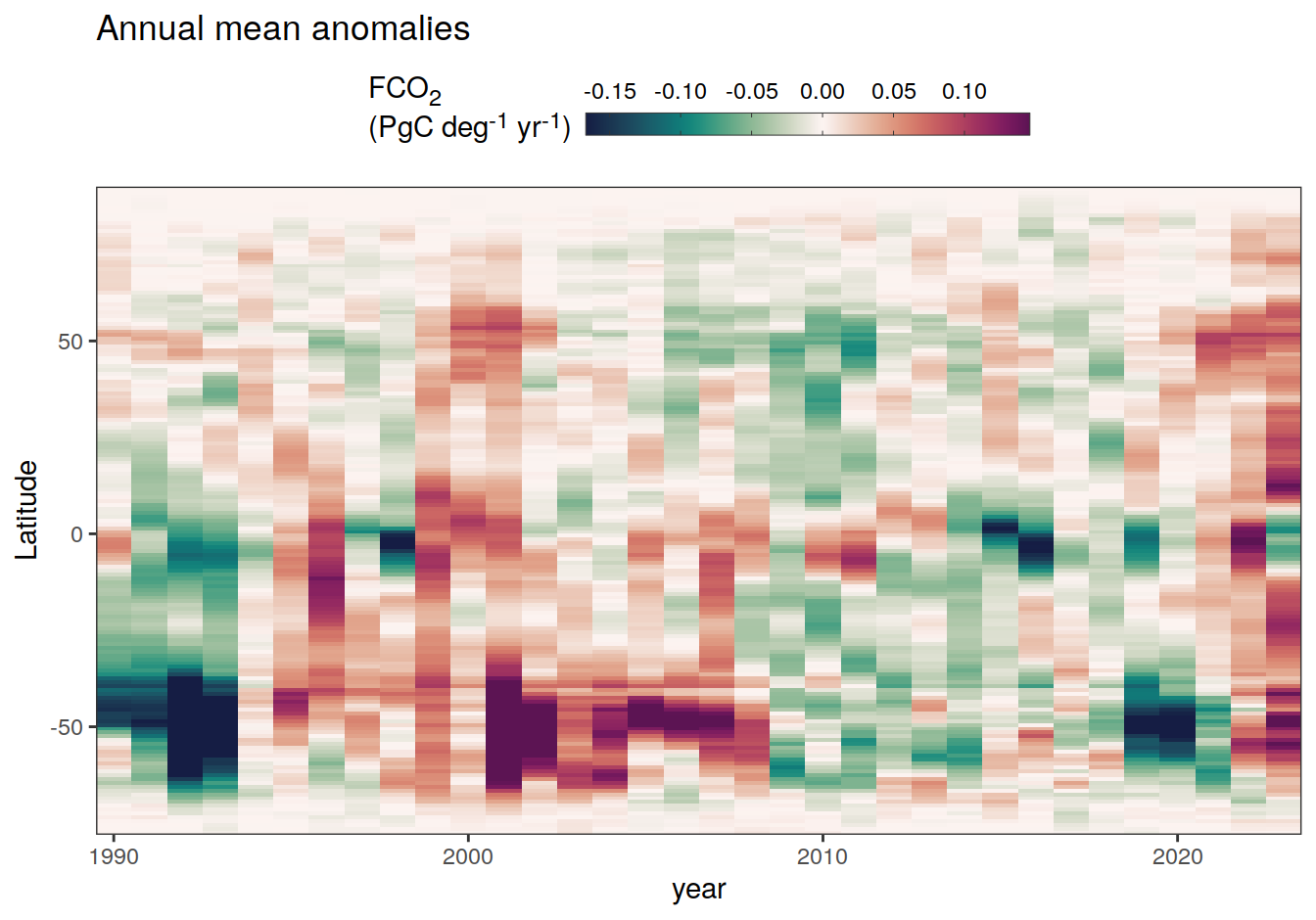
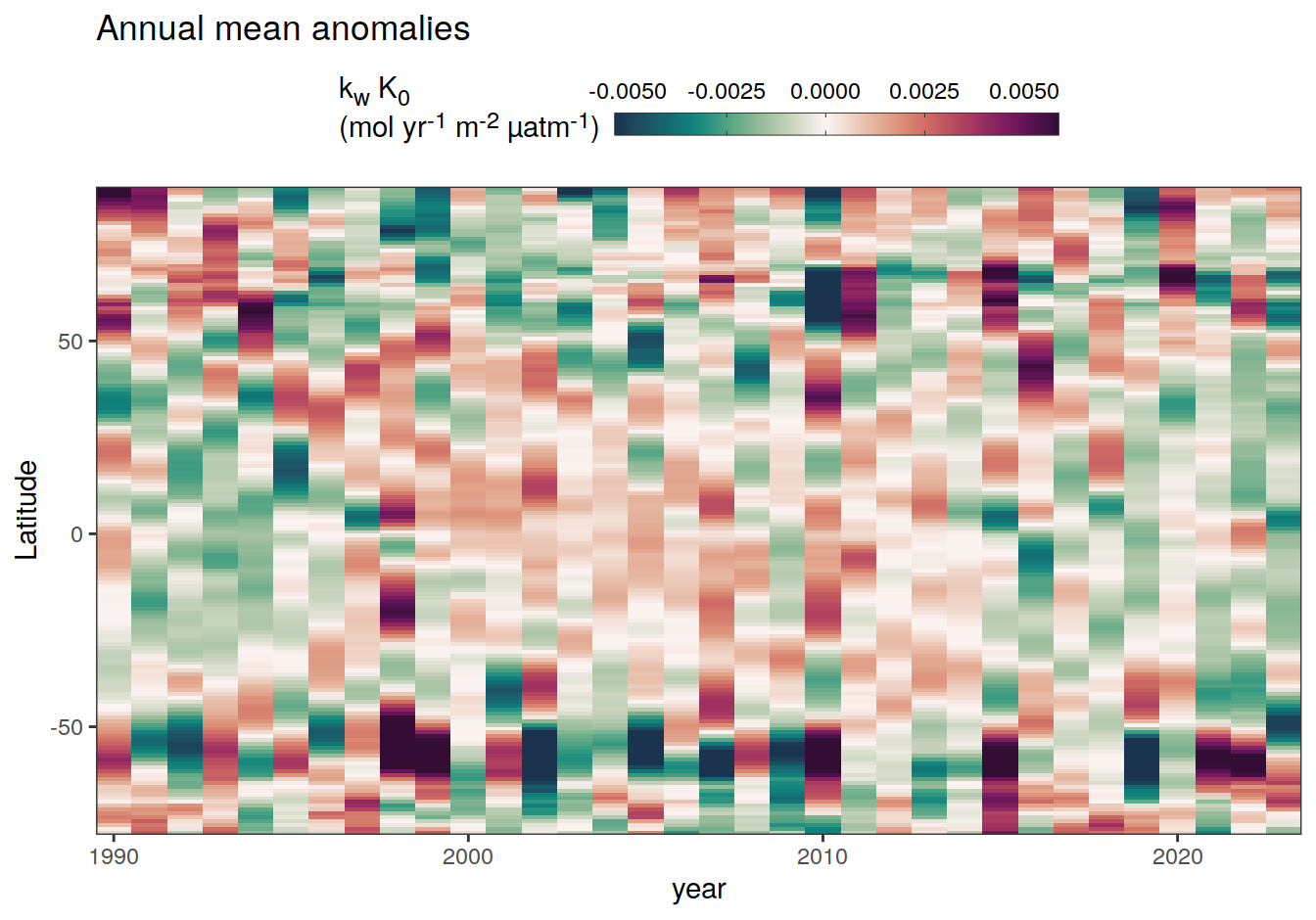
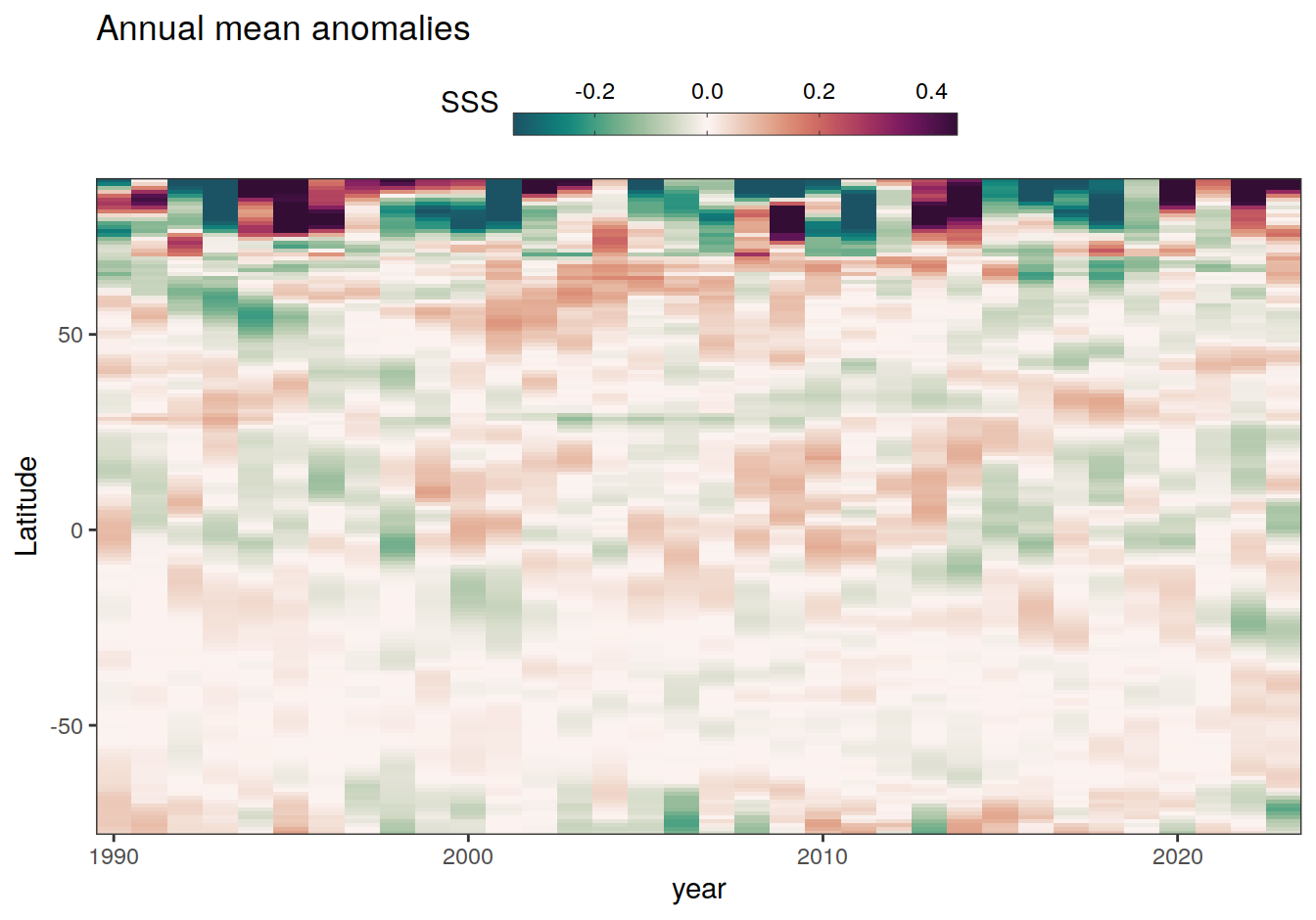
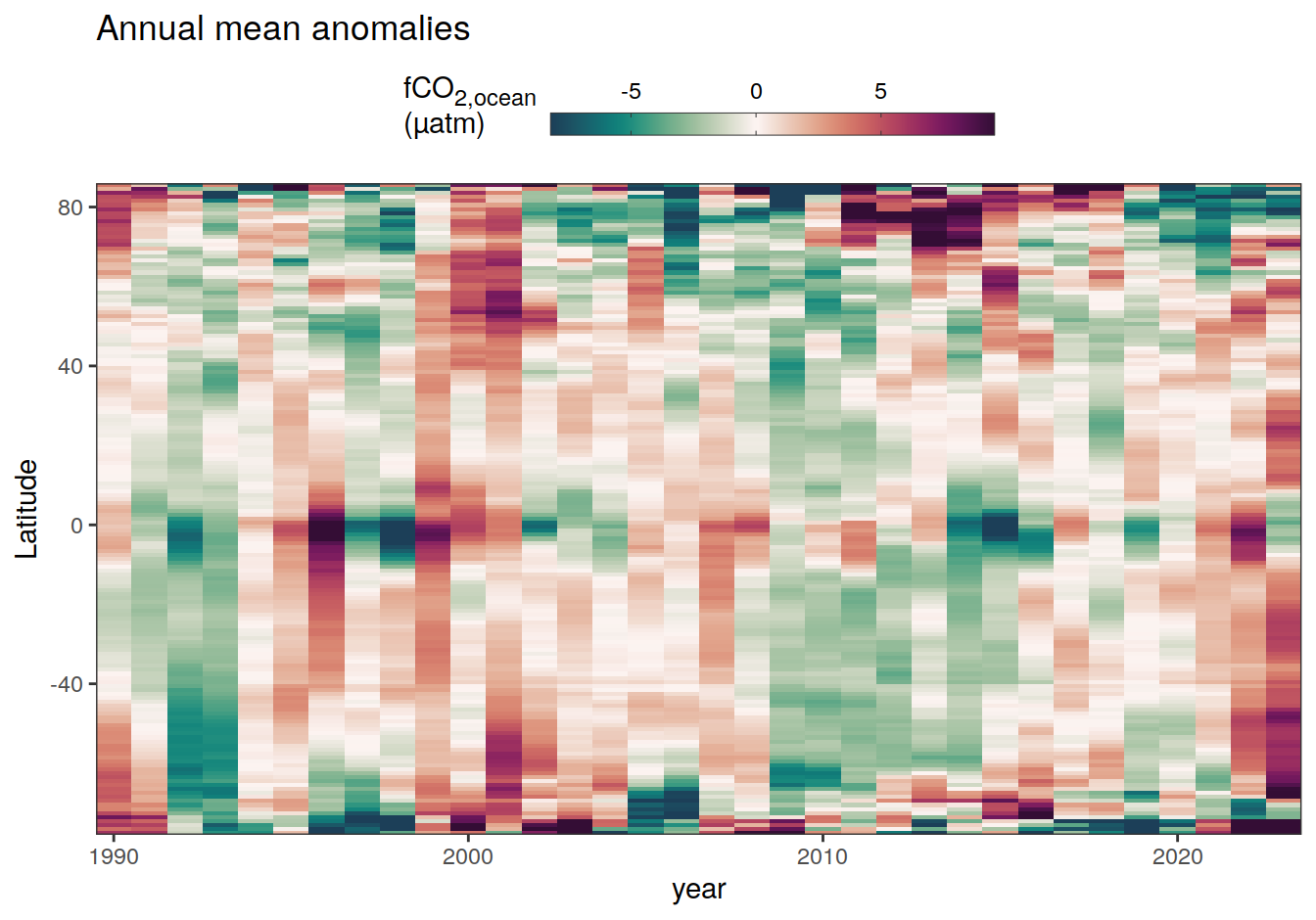
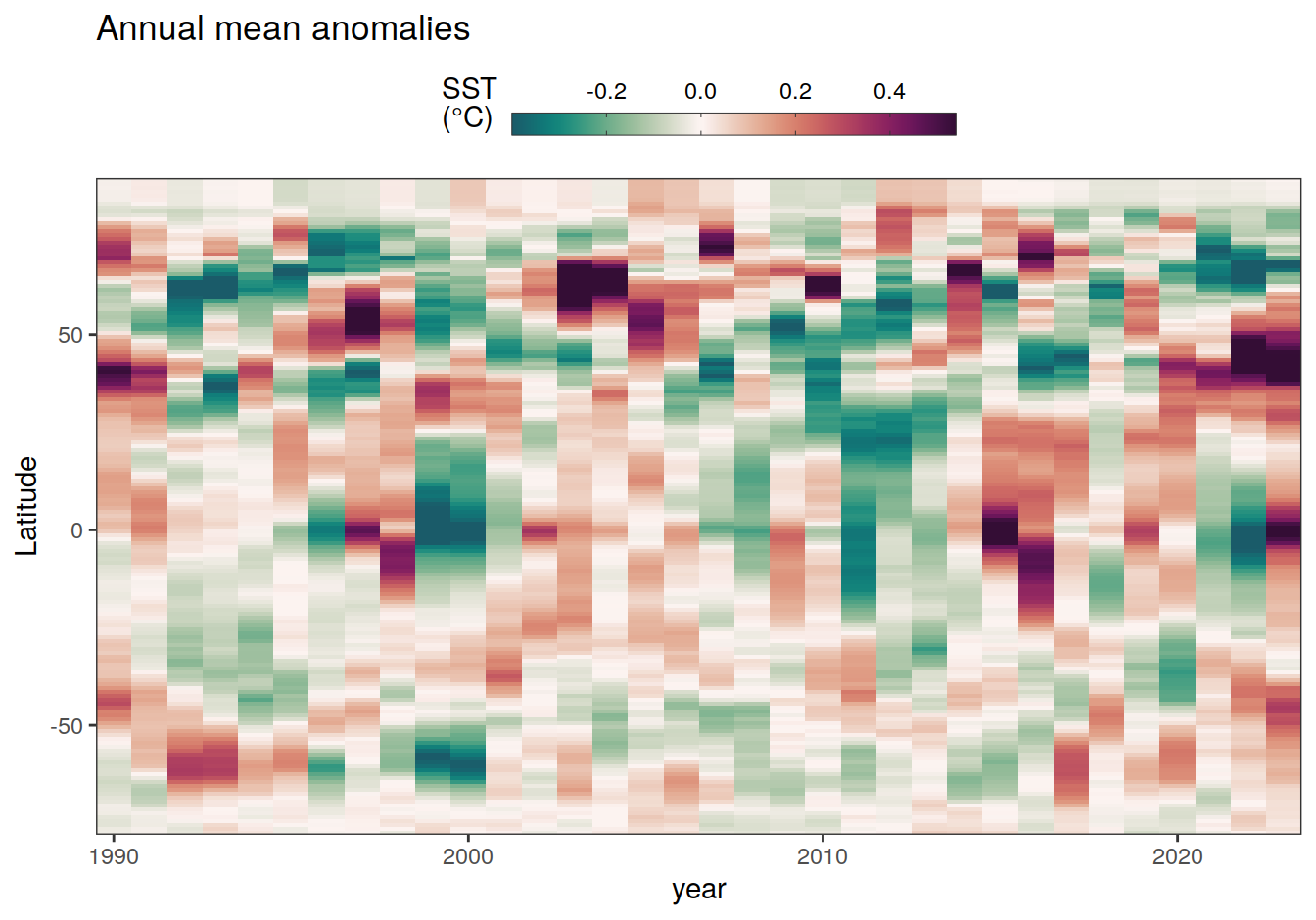
2023 annual anomalies
pco2_product_hovmoeller_annual_anomaly <-
pco2_product_hovmoeller_annual %>%
anomaly_determination(lat) %>%
filter(!is.na(resid))
pco2_product_hovmoeller_annual_anomaly %>%
# filter(name == "mld") %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x, aes(year, lat, fill = resid)) +
geom_raster() +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
coord_cartesian(expand = 0) +
labs(title = "Annual mean anomalies", y = "Latitude") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")
)
| Version | Author | Date |
|---|---|---|
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
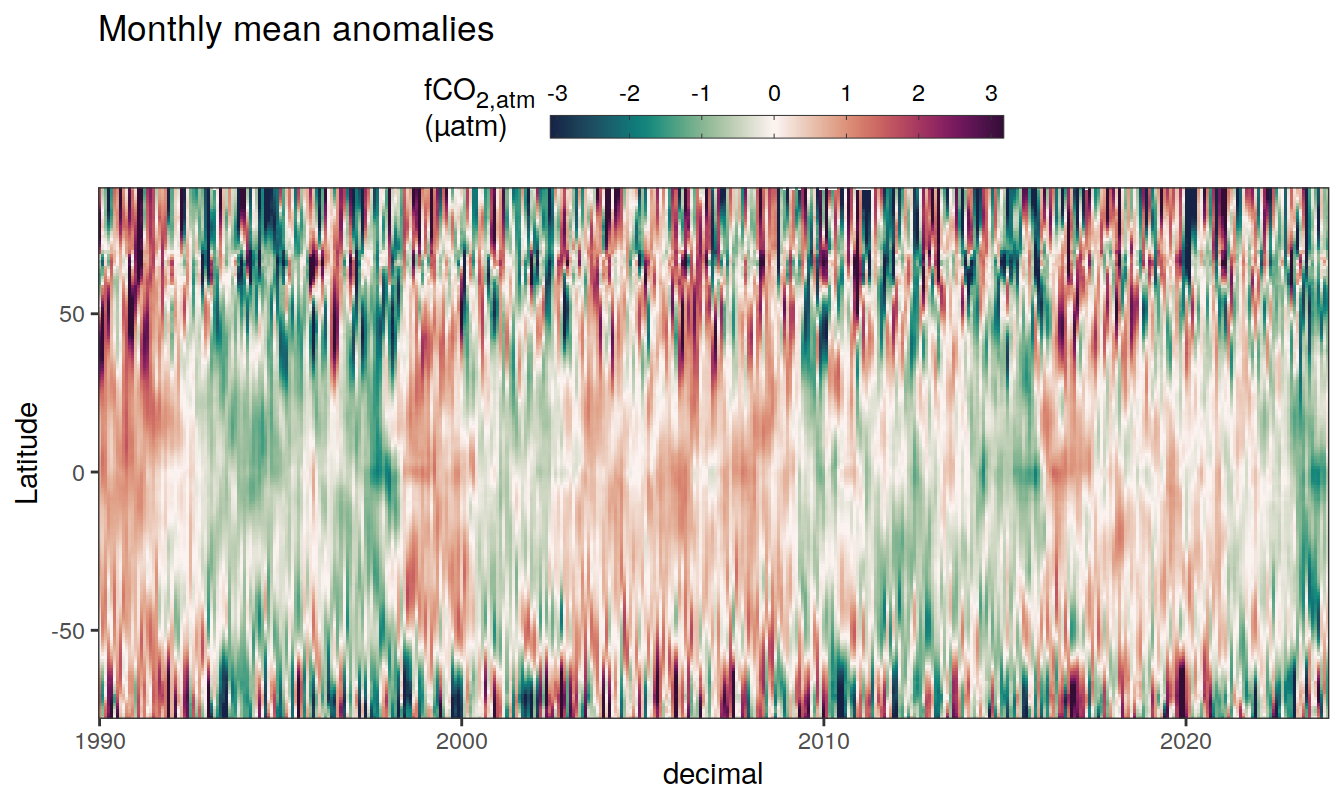
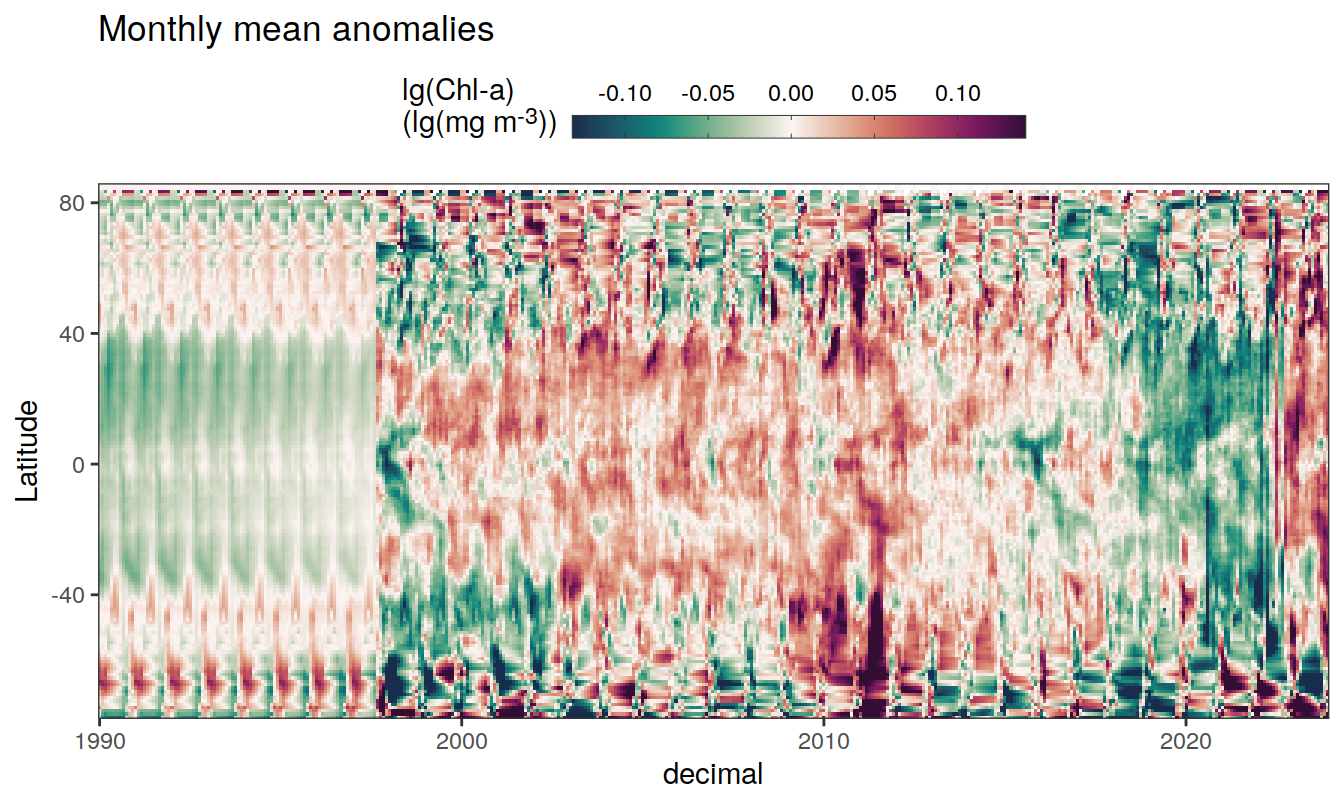
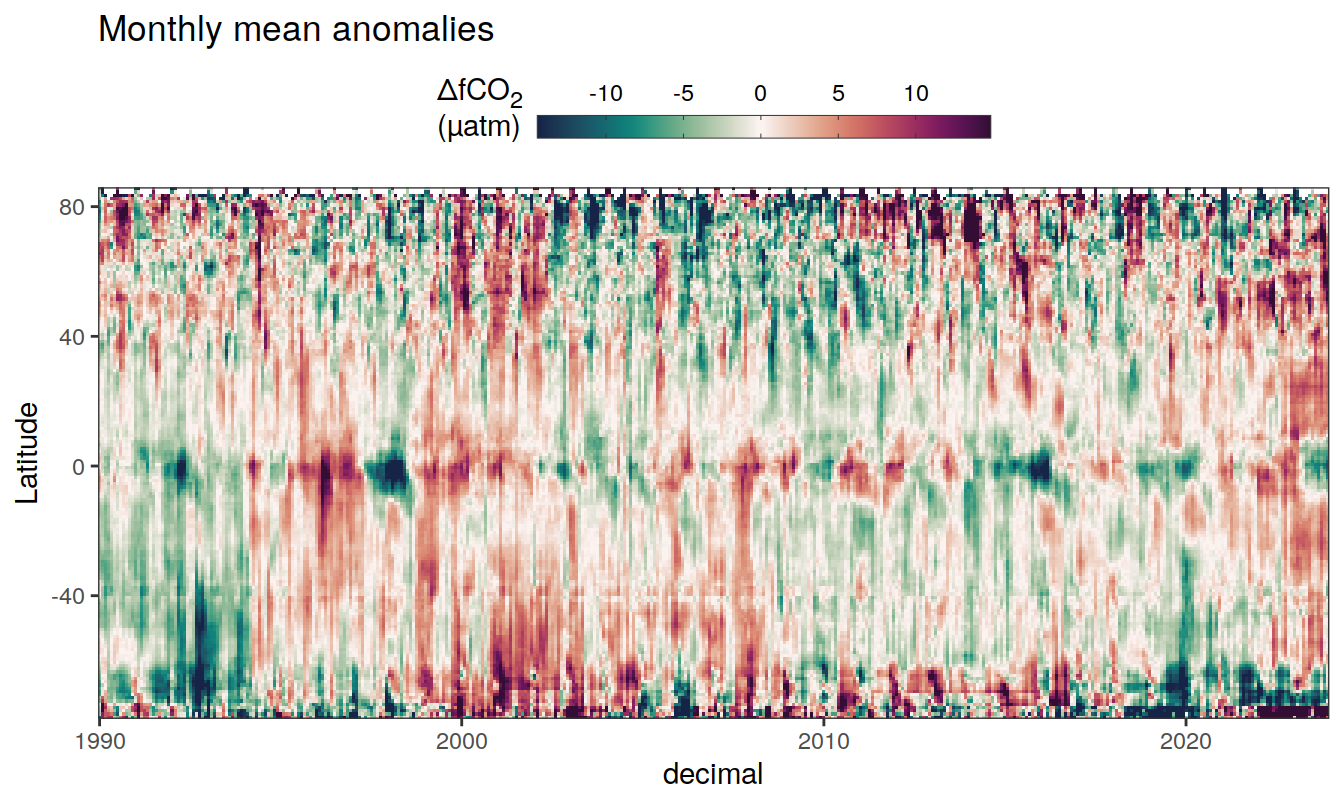
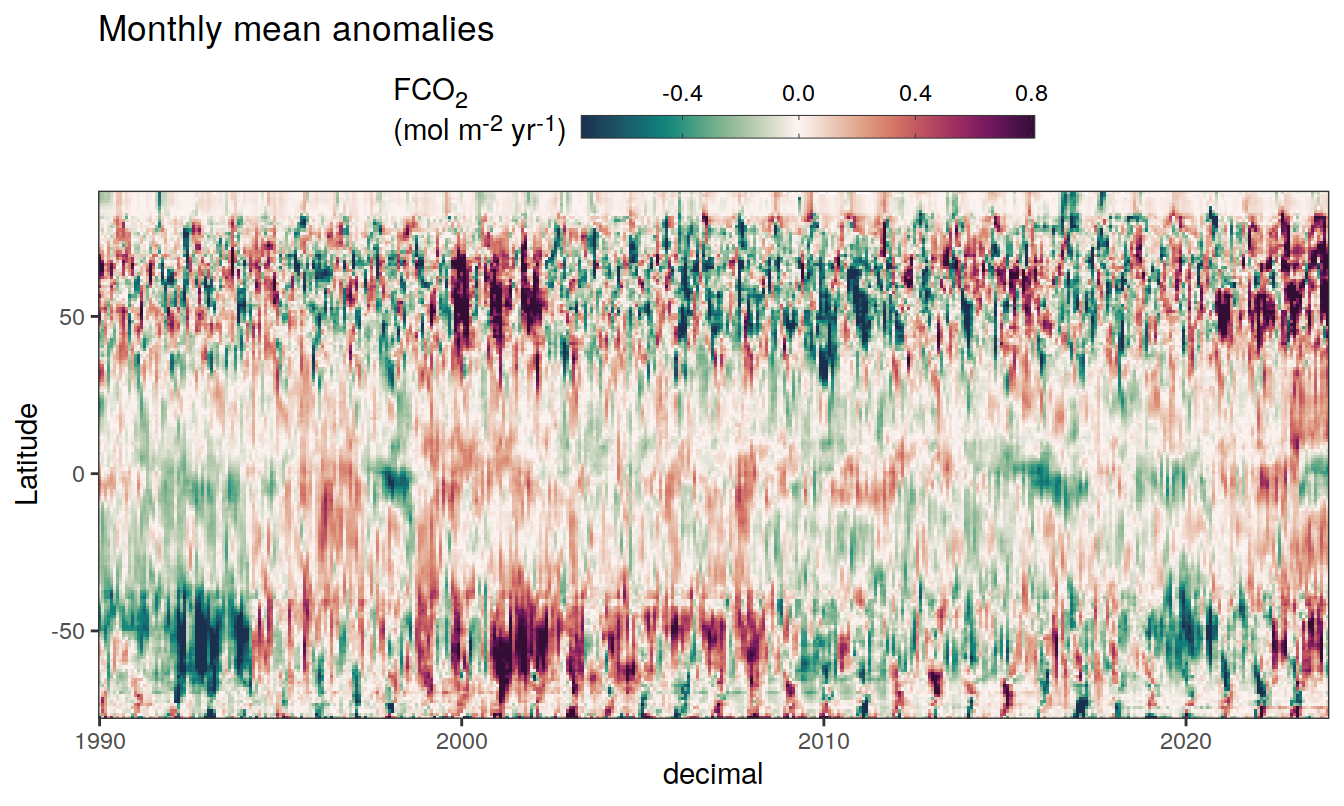
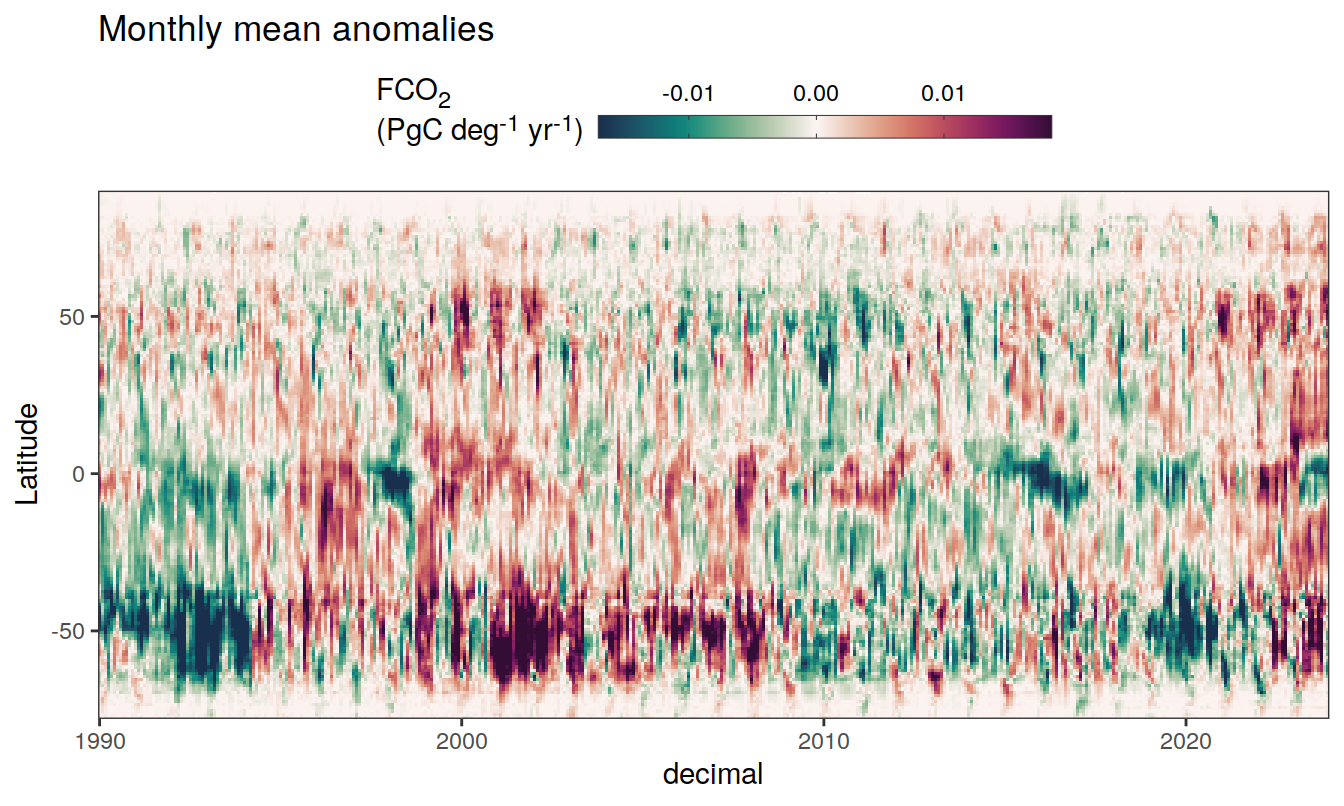
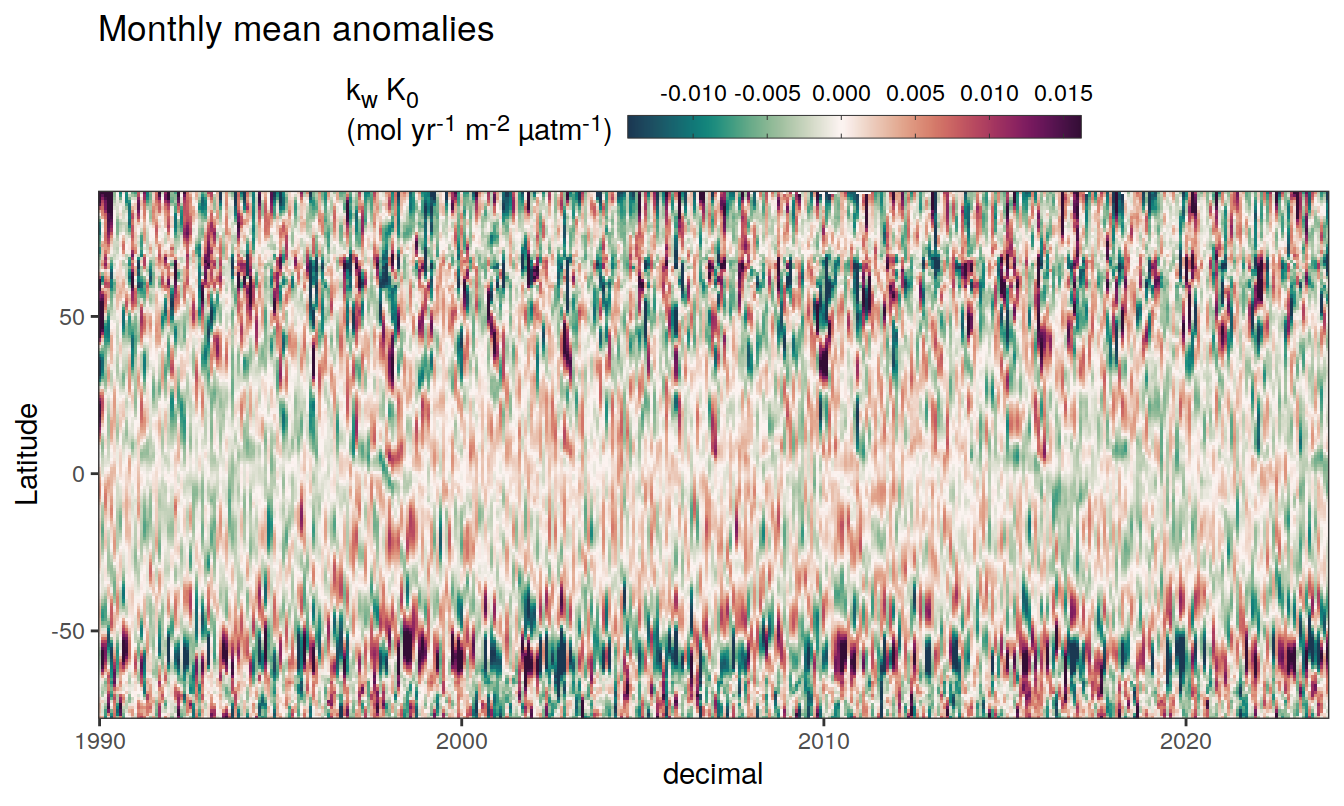
2023 monthly anomalies
pco2_product_hovmoeller_monthly_anomaly <-
pco2_product_hovmoeller_monthly %>%
select(-c(decimal)) %>%
anomaly_determination(lat, month) %>%
filter(!is.na(resid))
pco2_product_hovmoeller_monthly_anomaly <-
pco2_product_hovmoeller_monthly_anomaly %>%
mutate(decimal = year + (month - 1) / 12)
pco2_product_hovmoeller_monthly_anomaly %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(decimal, lat, fill = resid)) +
geom_raster() +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
coord_cartesian(expand = 0) +
labs(title = "Monthly mean anomalies",
y = "Latitude") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")
)
| Version | Author | Date |
|---|---|---|
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
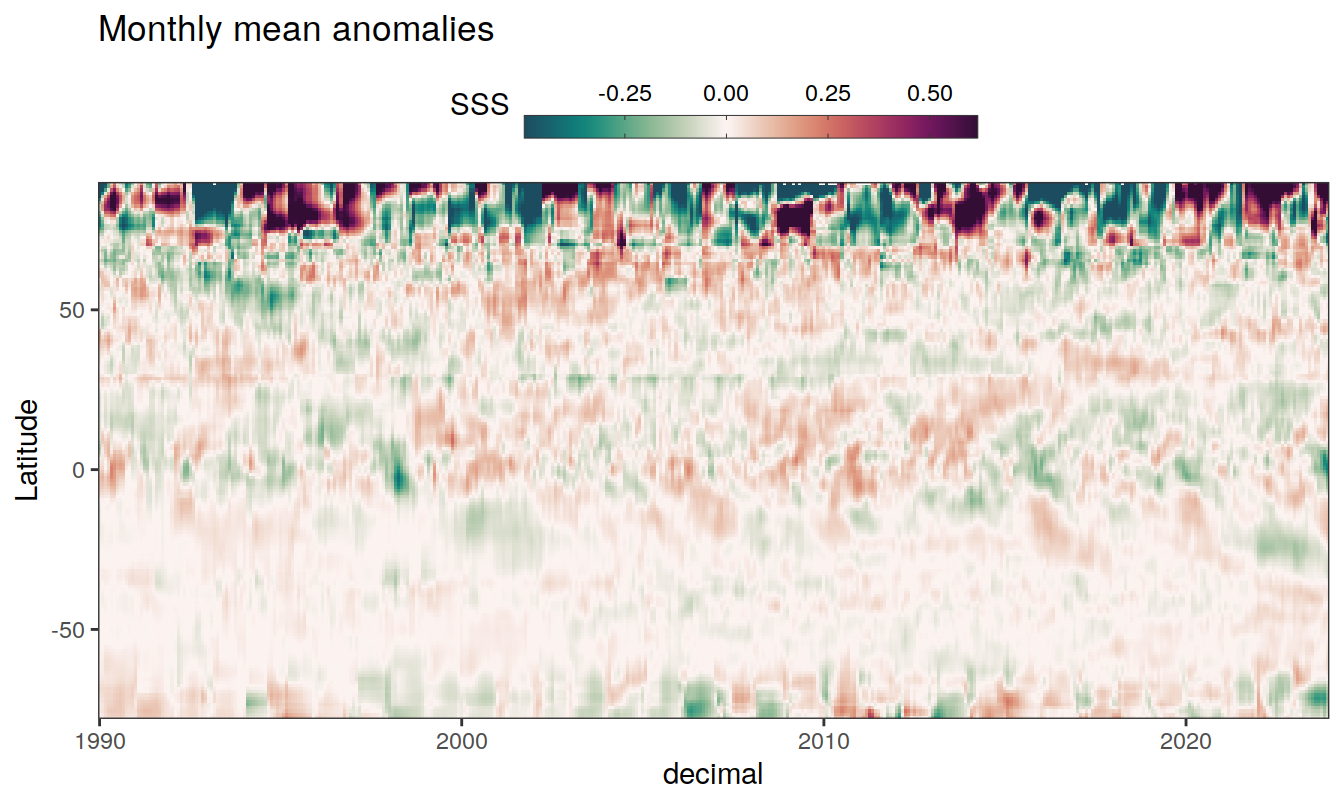
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
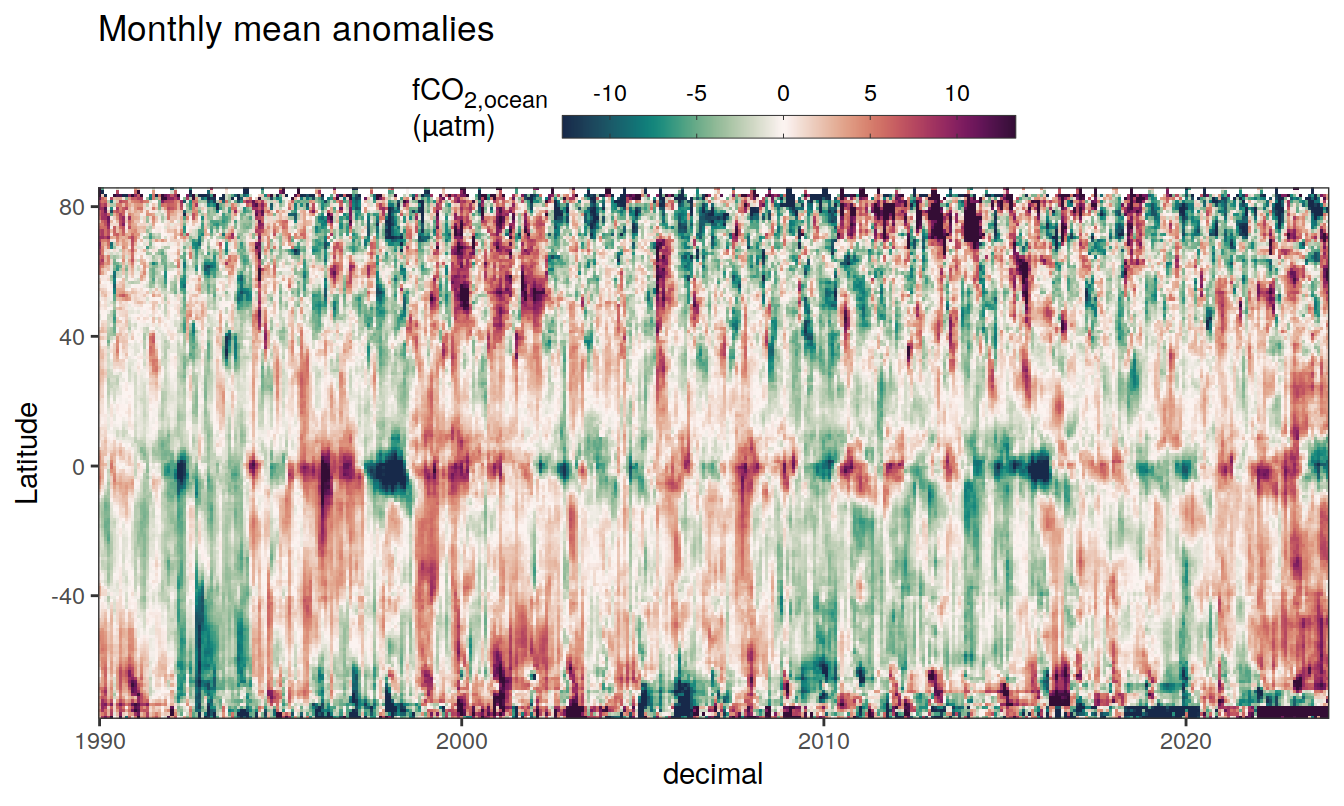
| Version | Author | Date |
|---|---|---|
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
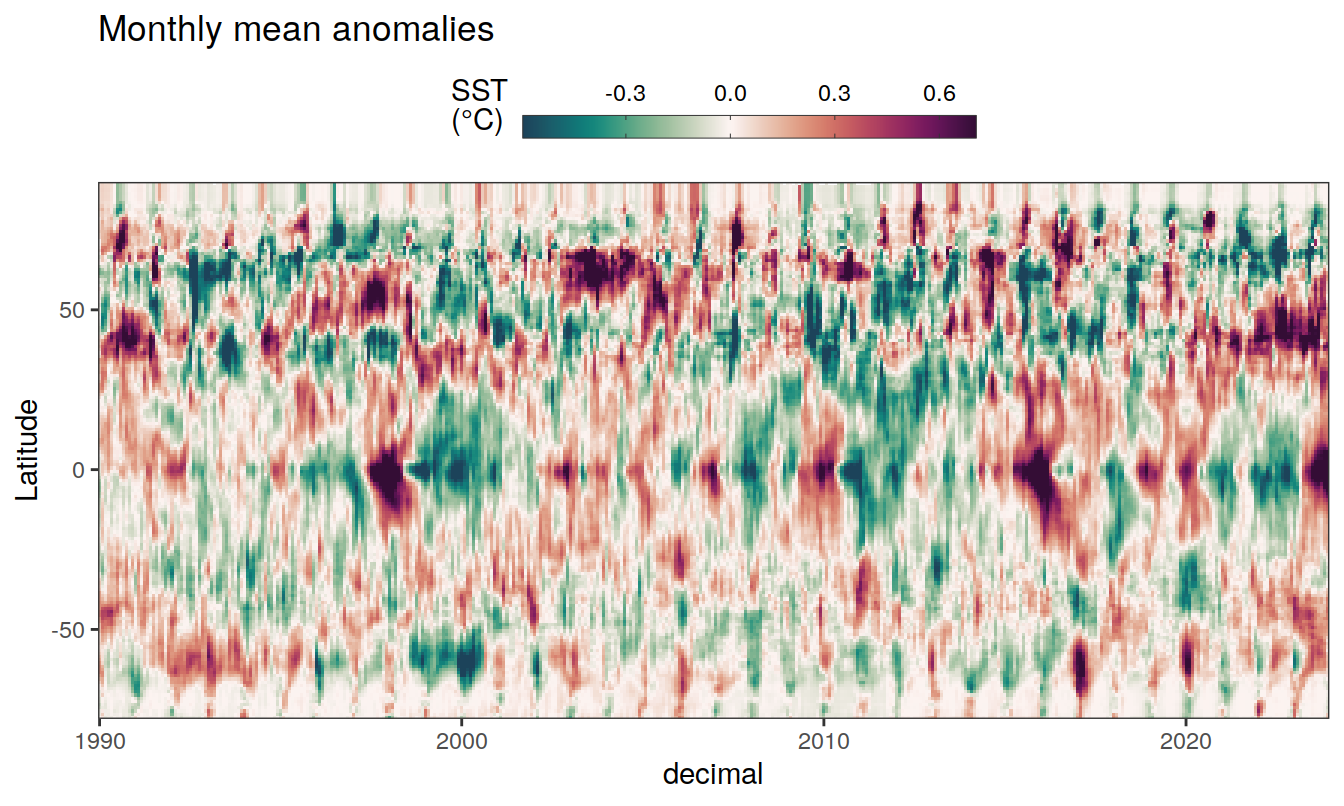
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
Three years prior 2023
pco2_product_hovmoeller_monthly_anomaly %>%
filter(between(year, 2023-2, 2023)) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(decimal, lat, fill = resid)) +
geom_raster() +
scale_fill_gradientn(
colours = cmocean("curl")(100),
rescaler = ~ scales::rescale_mid(.x, mid = 0),
name = labels_breaks(.x %>% distinct(name)),
limits = c(quantile(.x$resid, .01), quantile(.x$resid, .99)),
oob = squish
) +
coord_cartesian(expand = 0) +
labs(title = "Monthly mean anomalies",
y = "Latitude") +
guides(
fill = guide_colorbar(
barheight = unit(0.3, "cm"),
barwidth = unit(6, "cm"),
ticks = TRUE,
ticks.colour = "grey20",
frame.colour = "grey20",
label.position = "top",
direction = "horizontal"
)
) +
theme(legend.title = element_markdown(), legend.position = "top")
)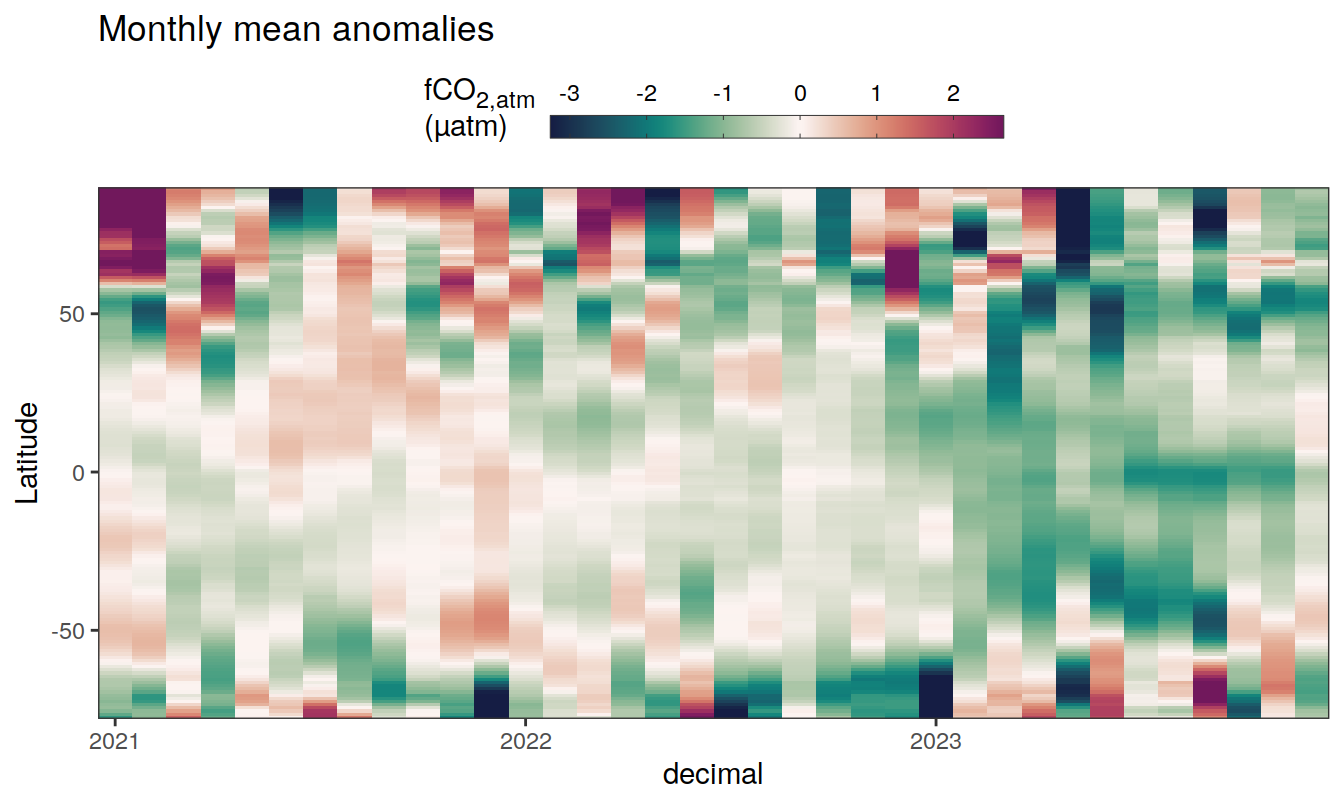
| Version | Author | Date |
|---|---|---|
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
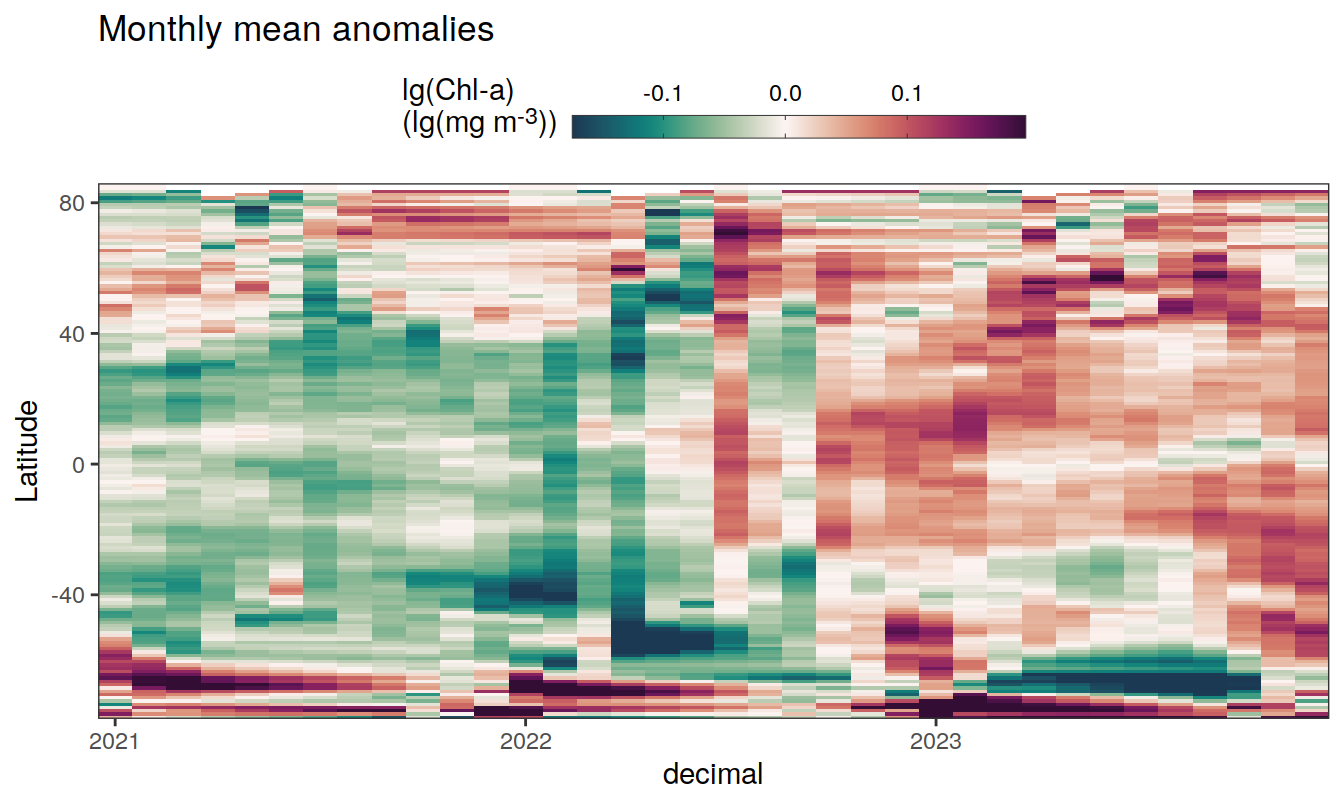
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
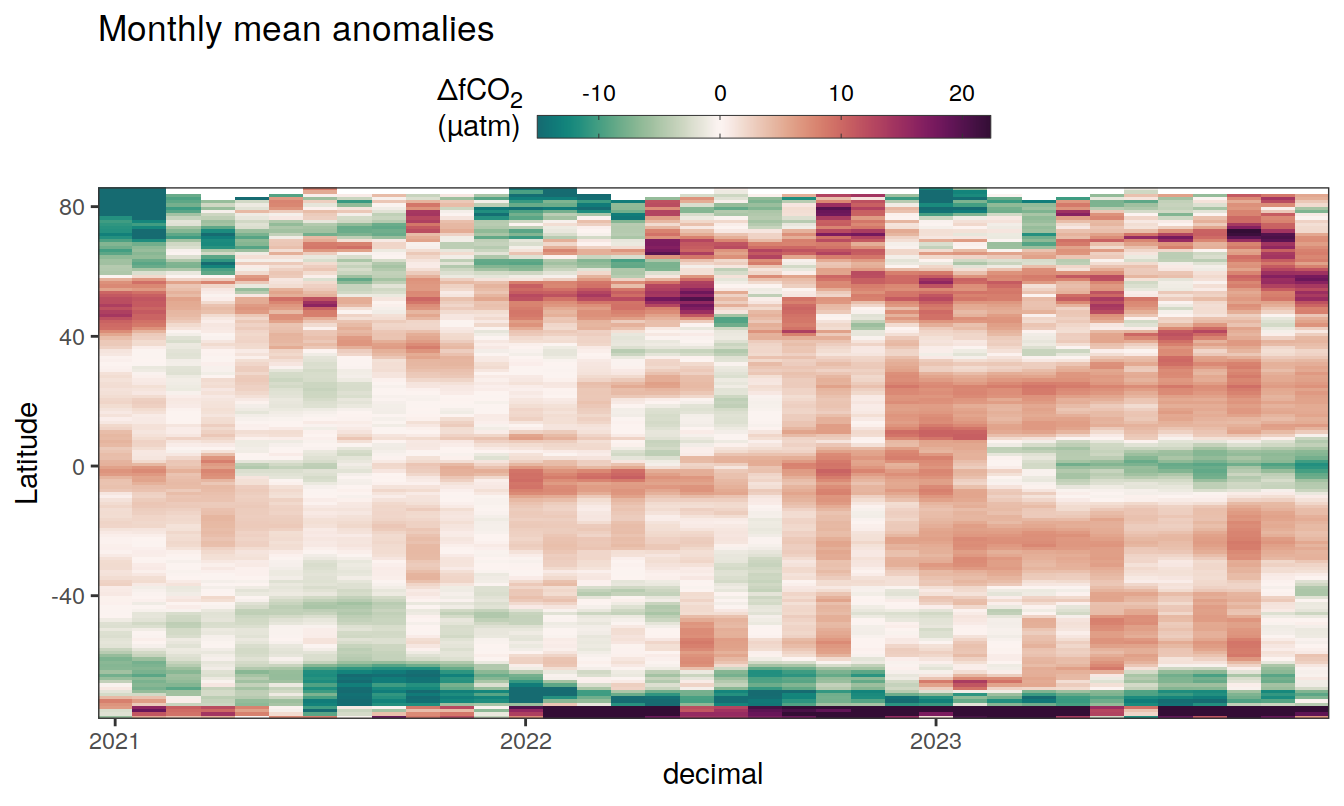
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
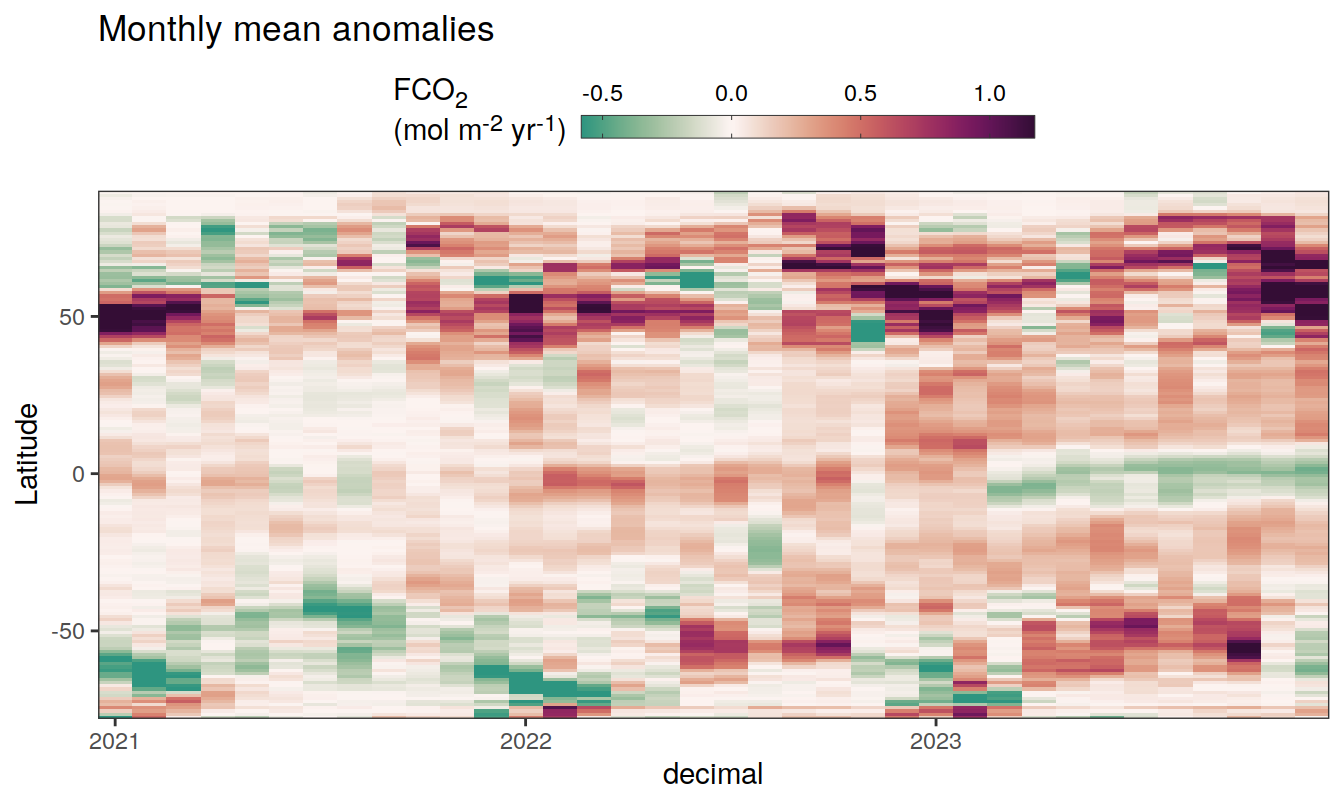
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
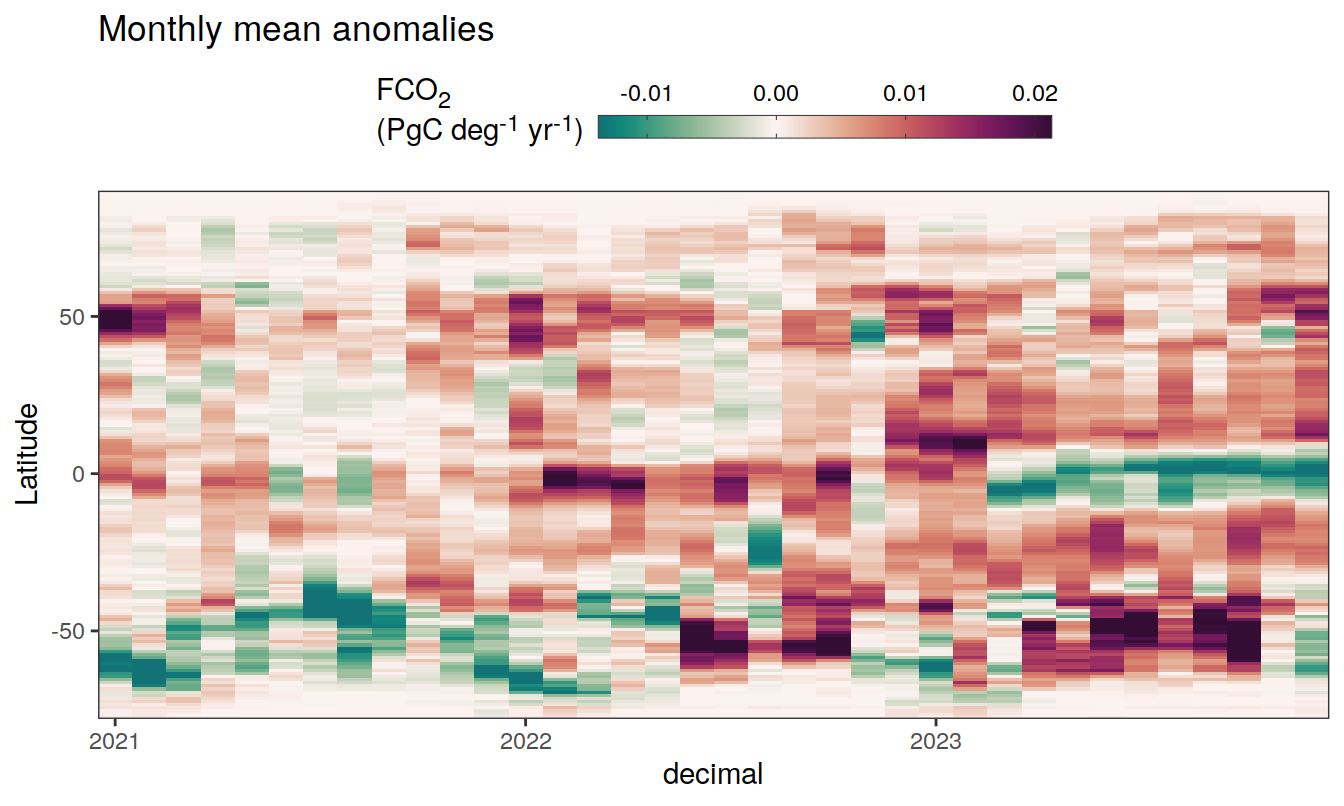
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
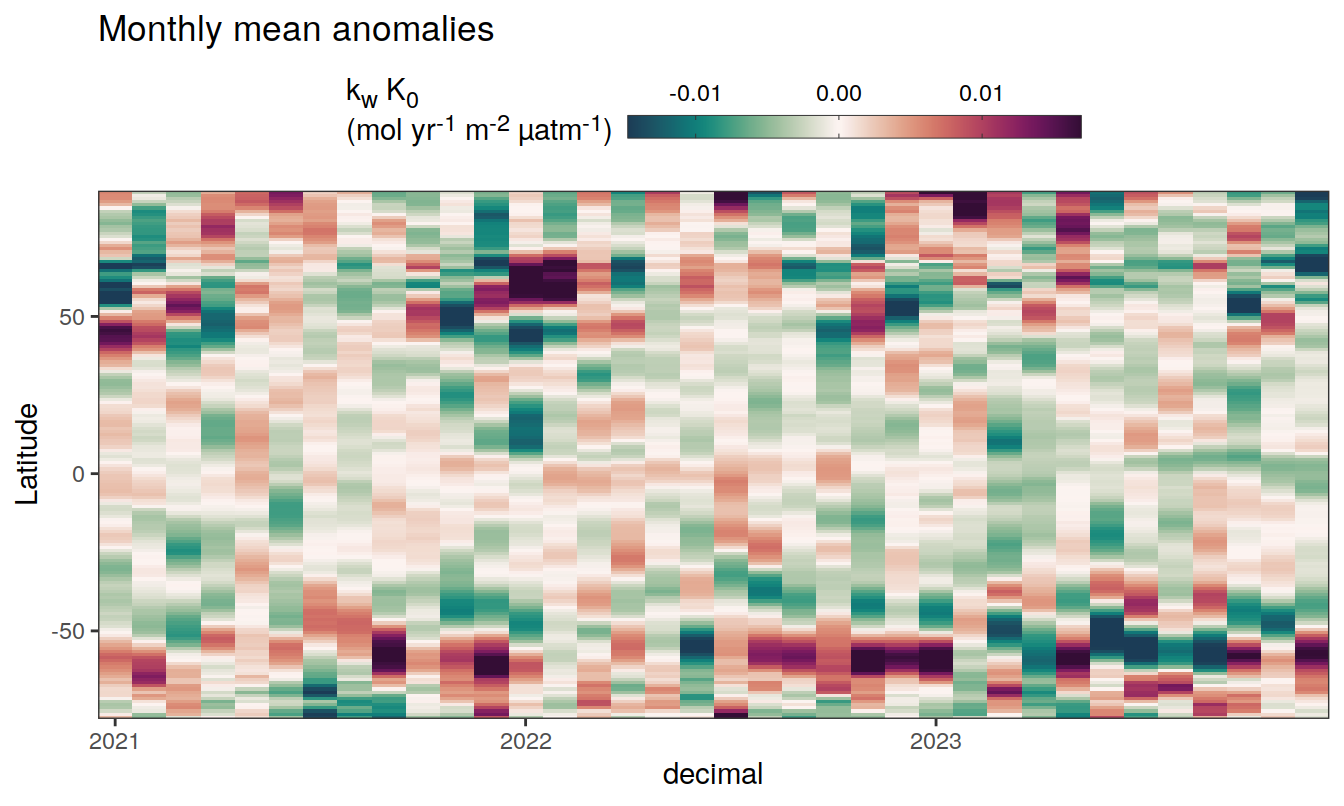
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
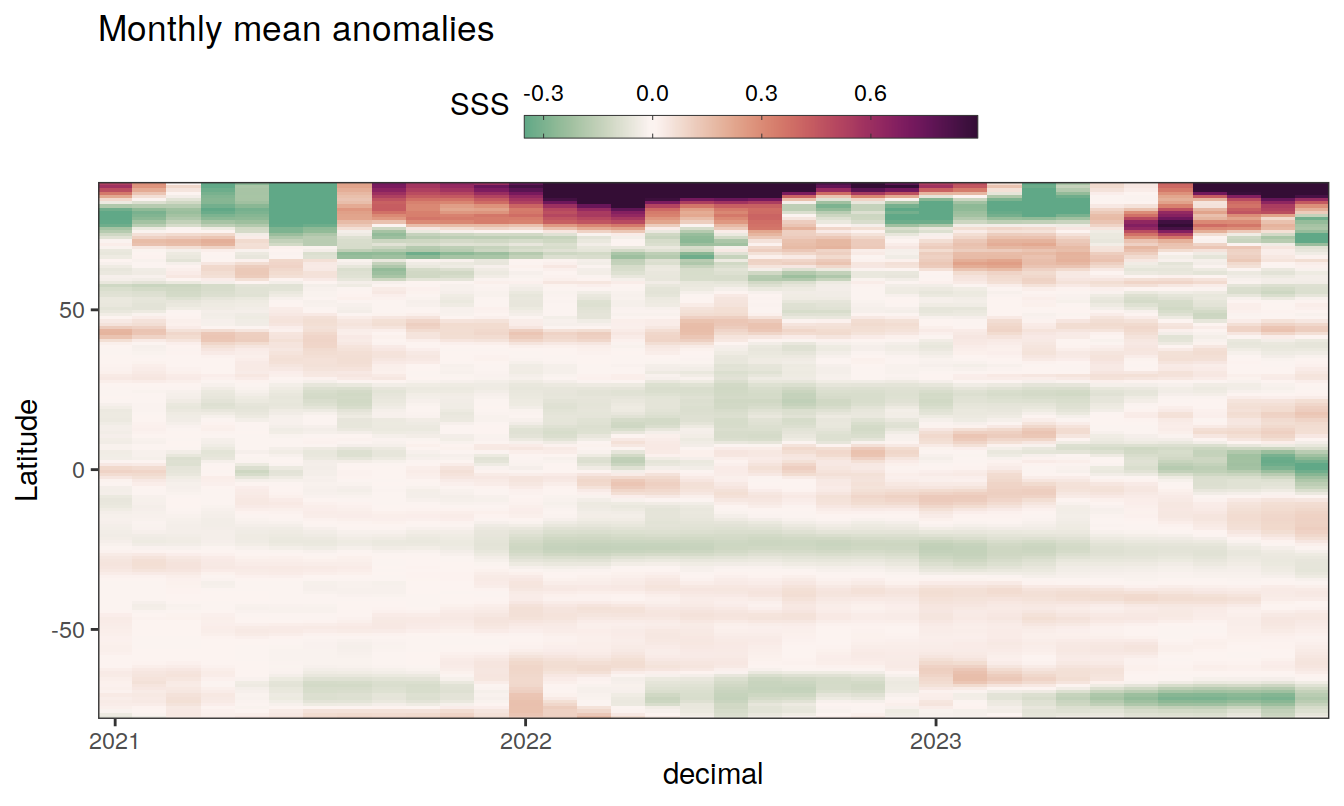
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
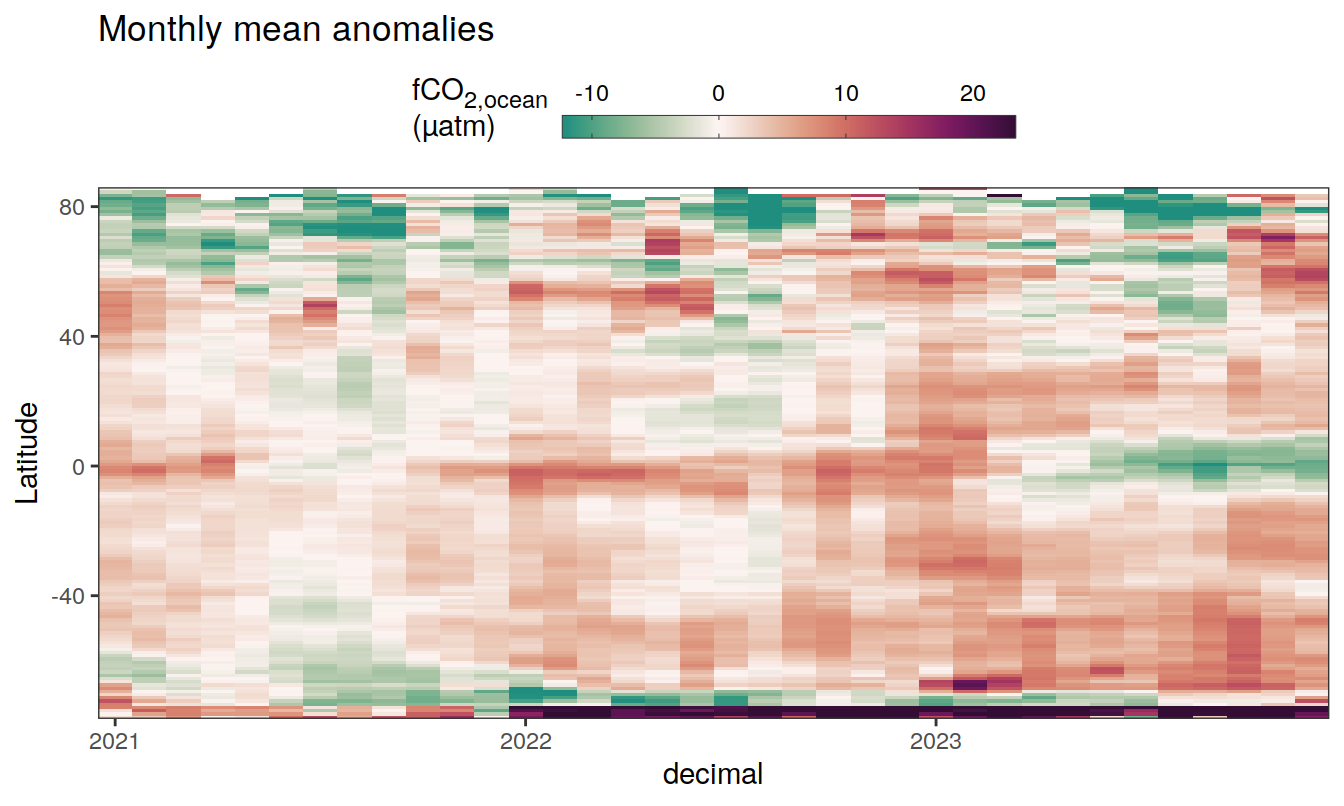
| Version | Author | Date |
|---|---|---|
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
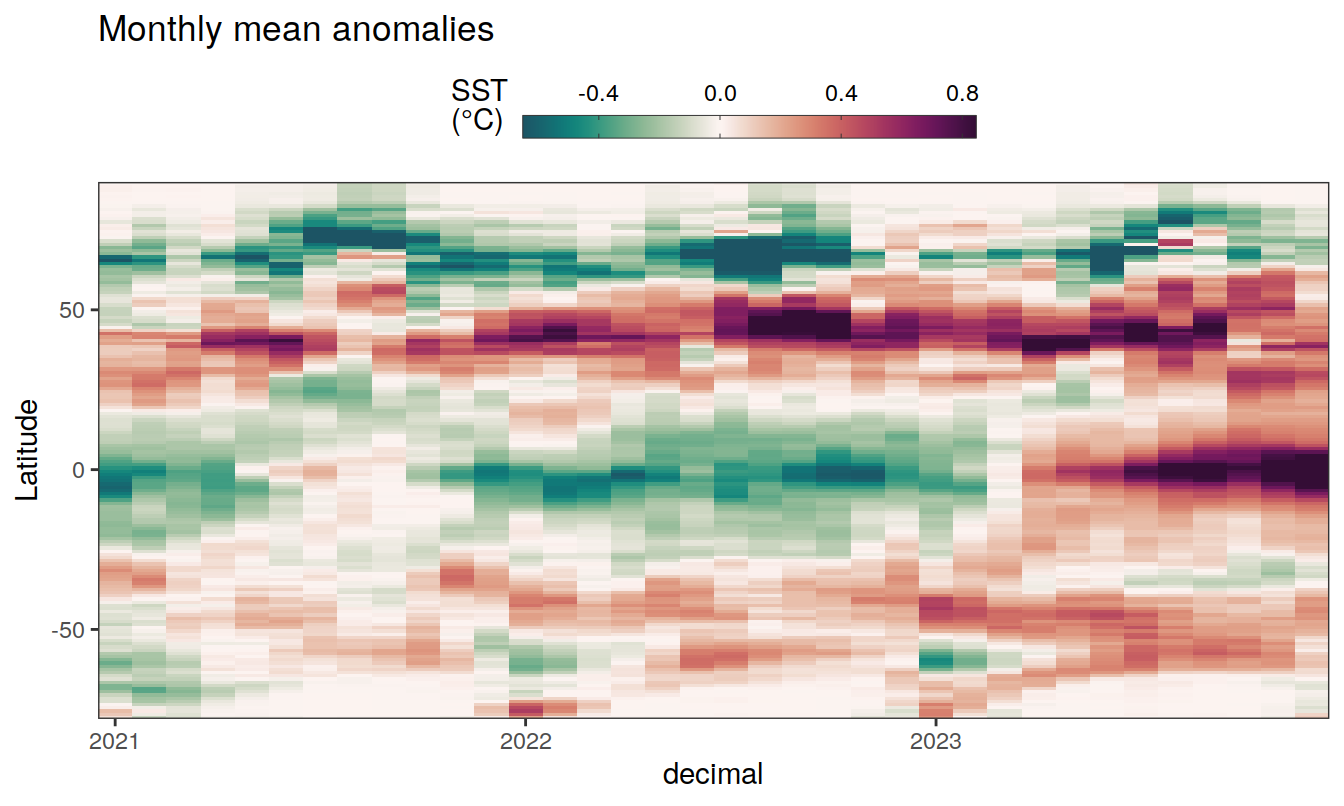
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_hovmoeller_monthly_anomaly %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_hovmoeller_monthly_anomaly.csv"
)
)
rm(
pco2_product_hovmoeller_annual,
pco2_product_hovmoeller_monthly,
pco2_product_hovmoeller_annual_anomaly,
pco2_product_hovmoeller_monthly_anomaly
)
gc()Regional means and integrals
The following plots show regionally averaged (or integrated) values of each variable as provided through the pCO2 product, as well as the anomalies from the prediction of a linear/quadratic fit.
Anomalies are first presented relative to the predicted annual mean of each year, hence preserving the seasonality. Furthermore, anomalies are presented relative to the predicted monthly mean values, such that the mean seasonality is removed.
2023 absolute values
Global non-polar
fig.height <- pco2_product_biome_monthly %>%
distinct(name) %>%
nrow()
fig.height <- (fig.height + 2) * 0.1pco2_product_biome_monthly %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(month, value, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Absolute values | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.title = element_blank(),
axis.title.y = element_blank()
)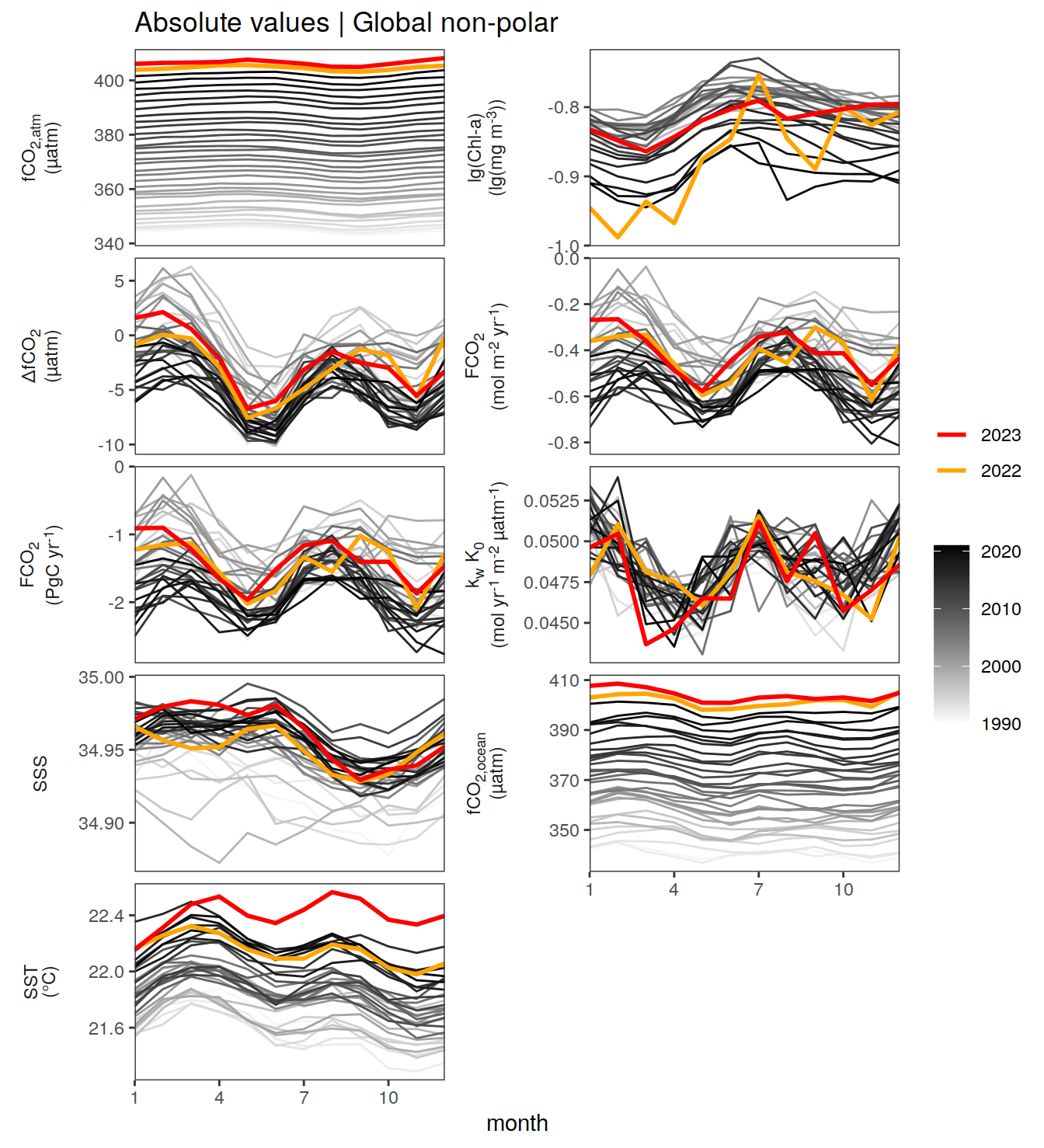
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
Key biomes
pco2_product_biome_monthly %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, value, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Absolute values | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.title = element_blank(),
axis.title.y = element_blank()
)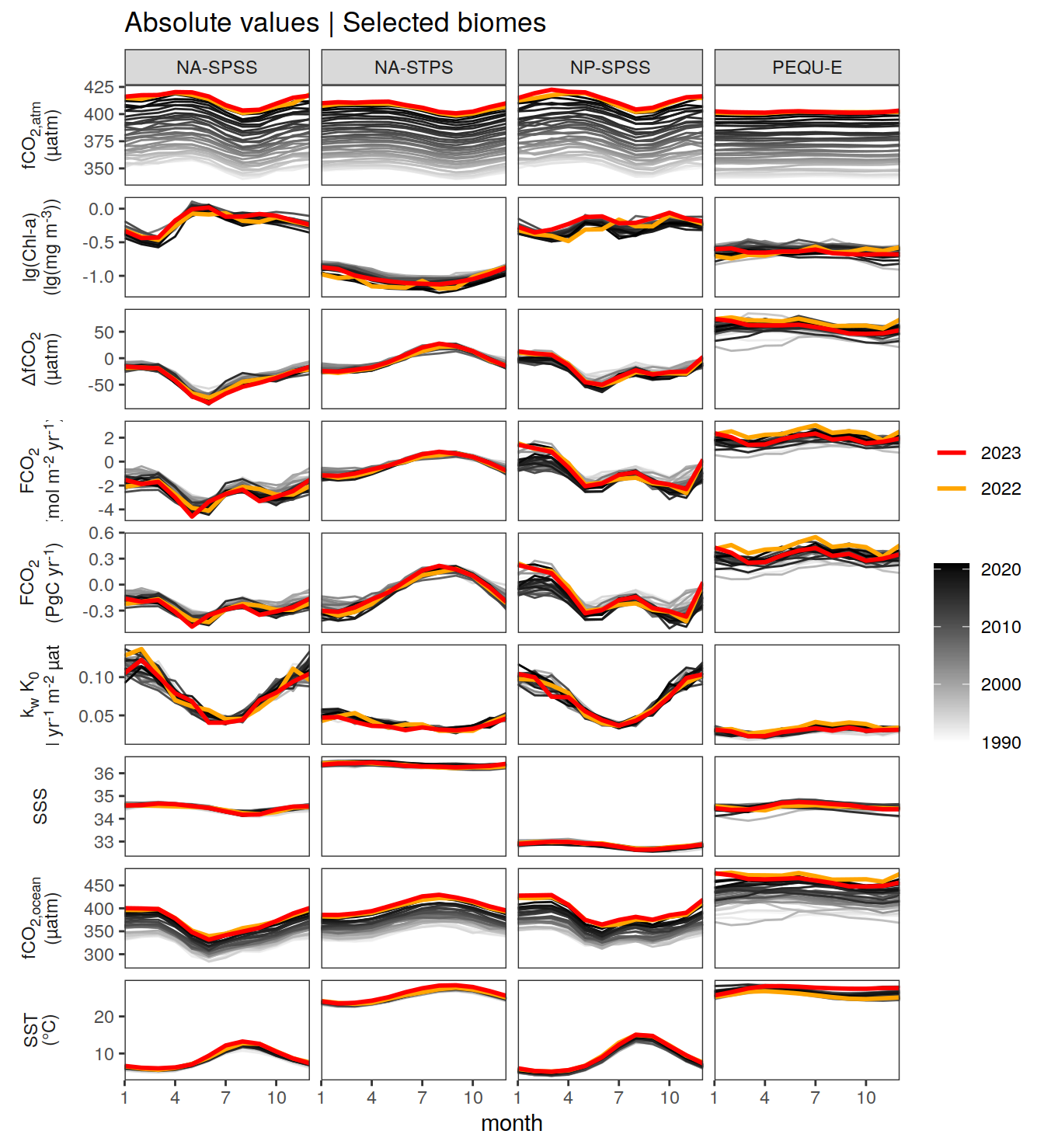
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_monthly %>%
filter(biome %in% key_biomes) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(month, value, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(
data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1
) +
scale_color_manual(
values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = paste("Absolute values |", .x$biome)) +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
legend.title = element_blank(),
axis.title.y = element_blank()
)
)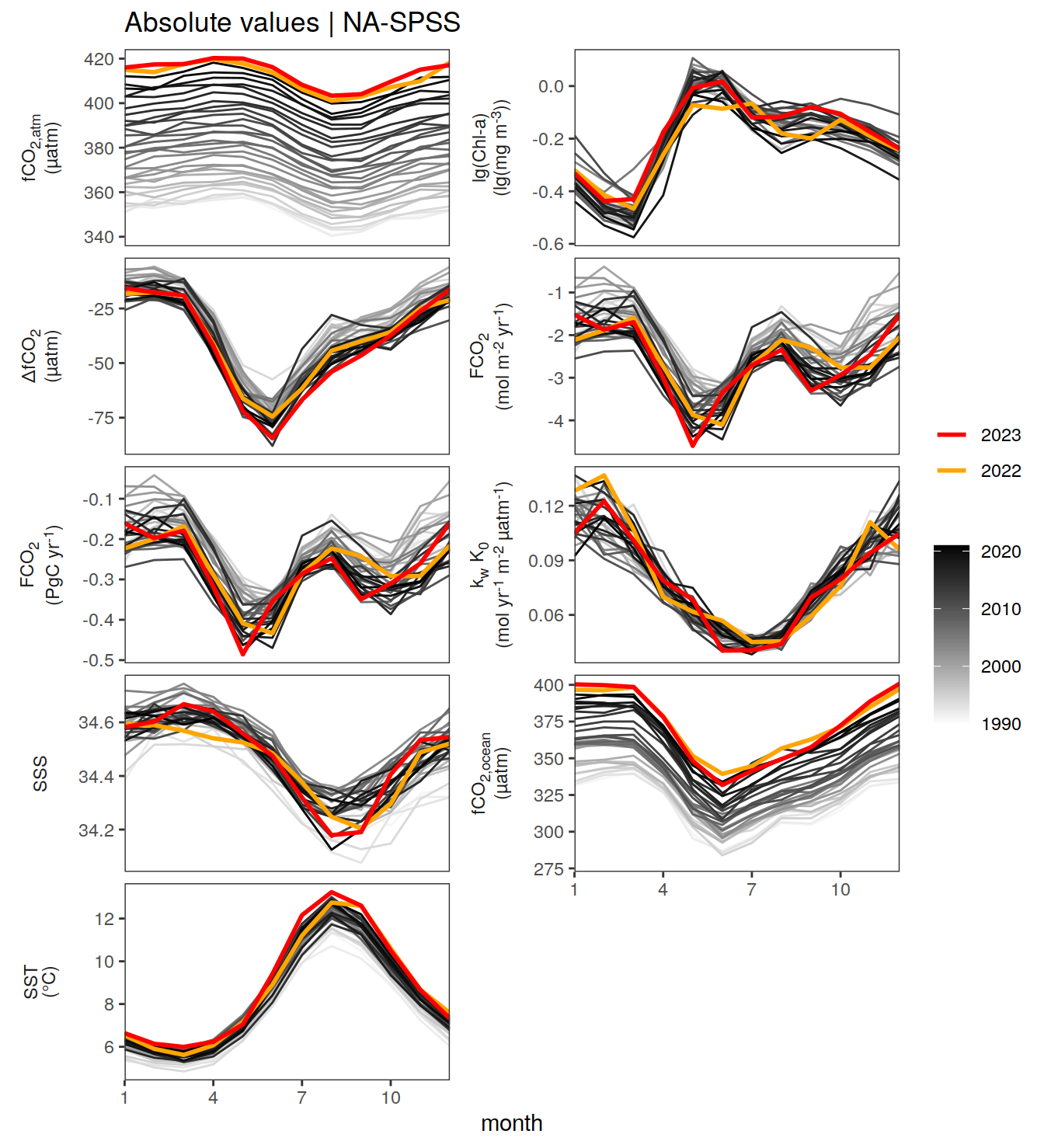
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
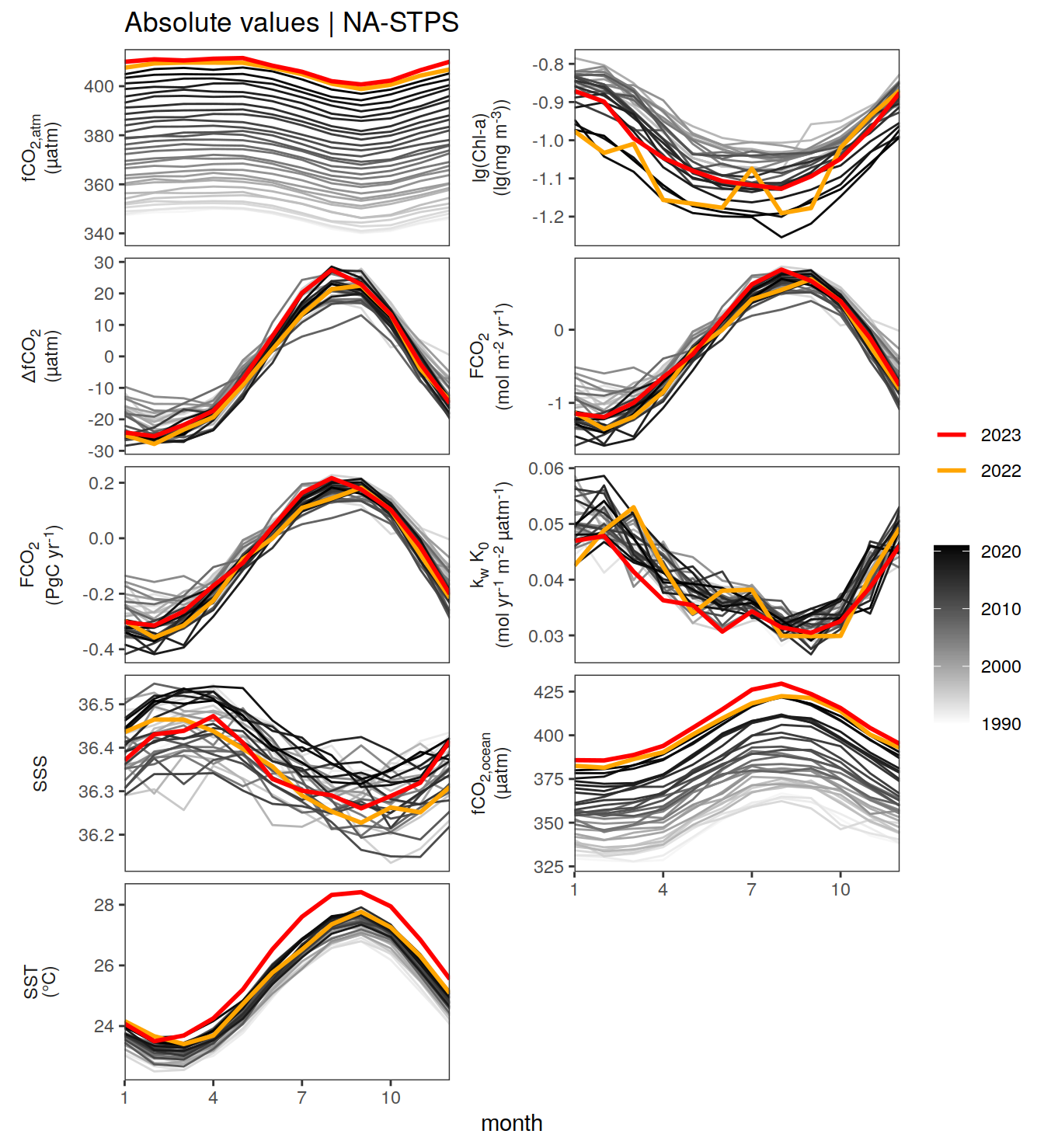
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
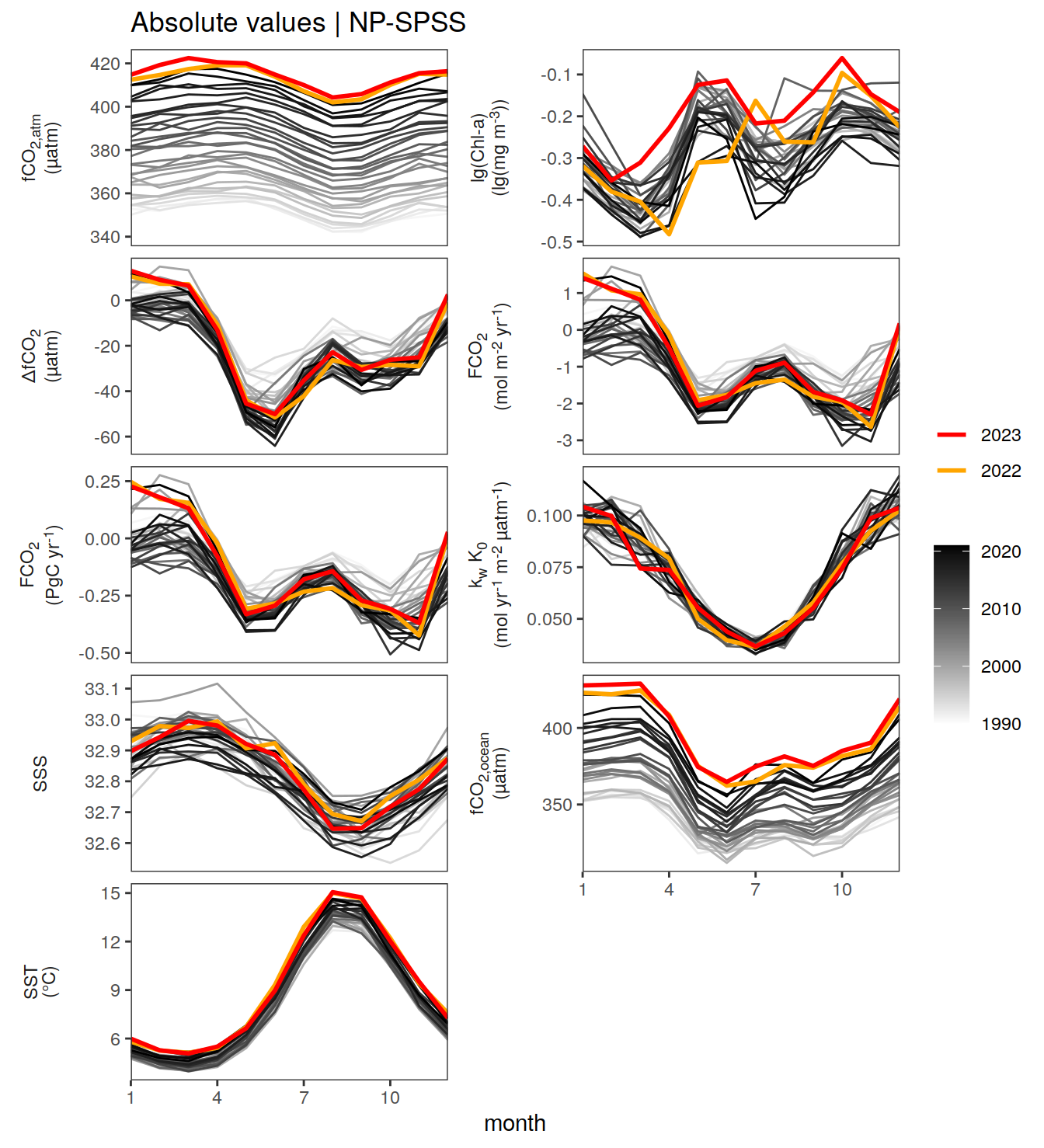
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
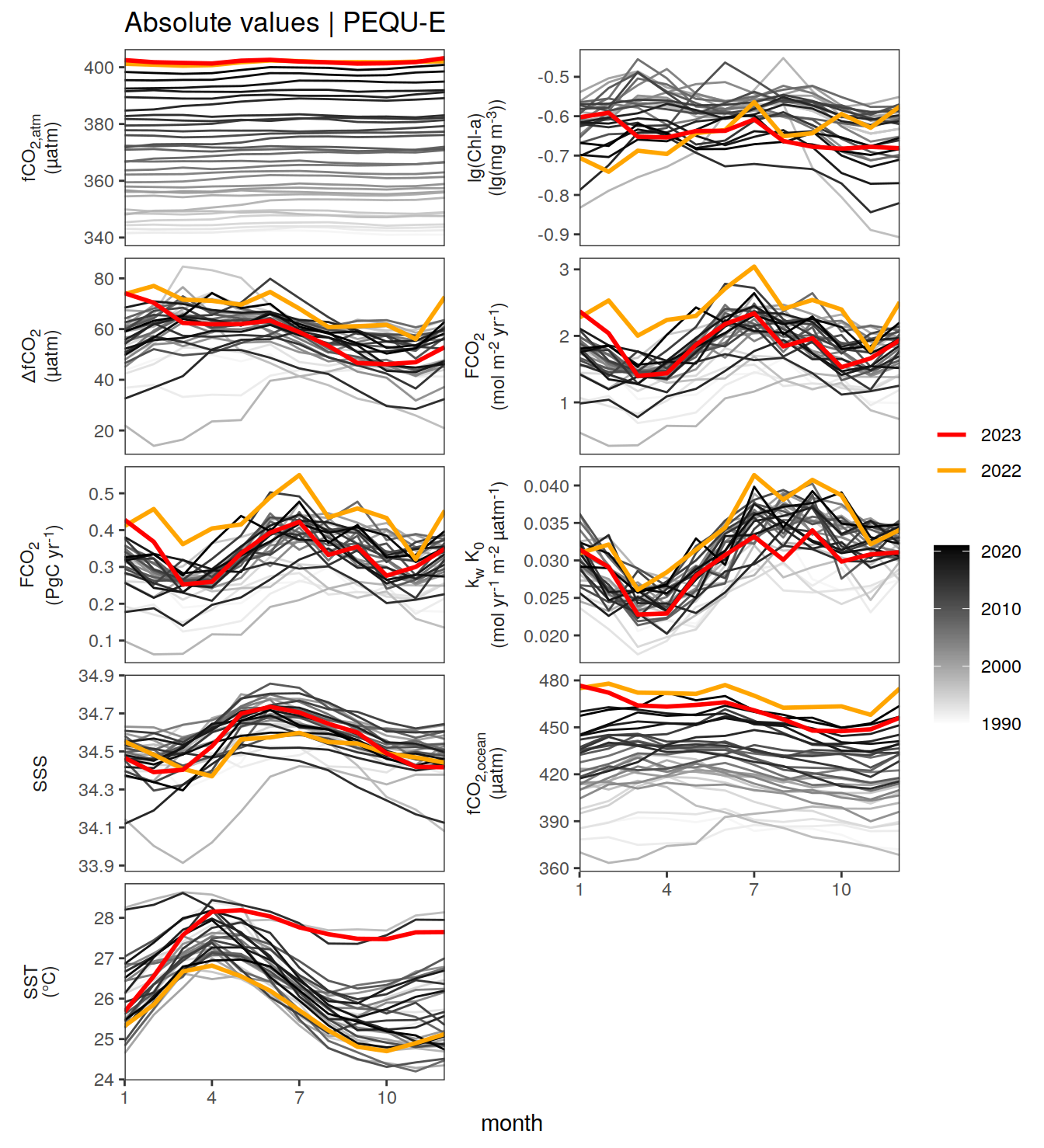
| Version | Author | Date |
|---|---|---|
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
2023 anomalies
Annual mean trends
pco2_product_biome_annual %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(year, value)) +
geom_path() +
geom_point(data = . %>% filter(year != 2023)) +
geom_point(data = . %>% filter(year == 2023),
shape = 1) +
geom_smooth(data = . %>% filter(year != 2023,
!(name %in% name_quadratic_fit)),
method = "lm",
fullrange = TRUE,
aes(col = "linear"),
se = FALSE) +
geom_smooth(data = . %>% filter(year != 2023,
name %in% name_quadratic_fit),
method = "lm",
fullrange = TRUE,
formula = y ~ poly(x,2),
aes(col = "poly2"),
se = FALSE) +
scale_color_brewer(
palette = "Set1",
name = paste("Regression fit\nexcl.", 2023)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Annual mean trends | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank()
)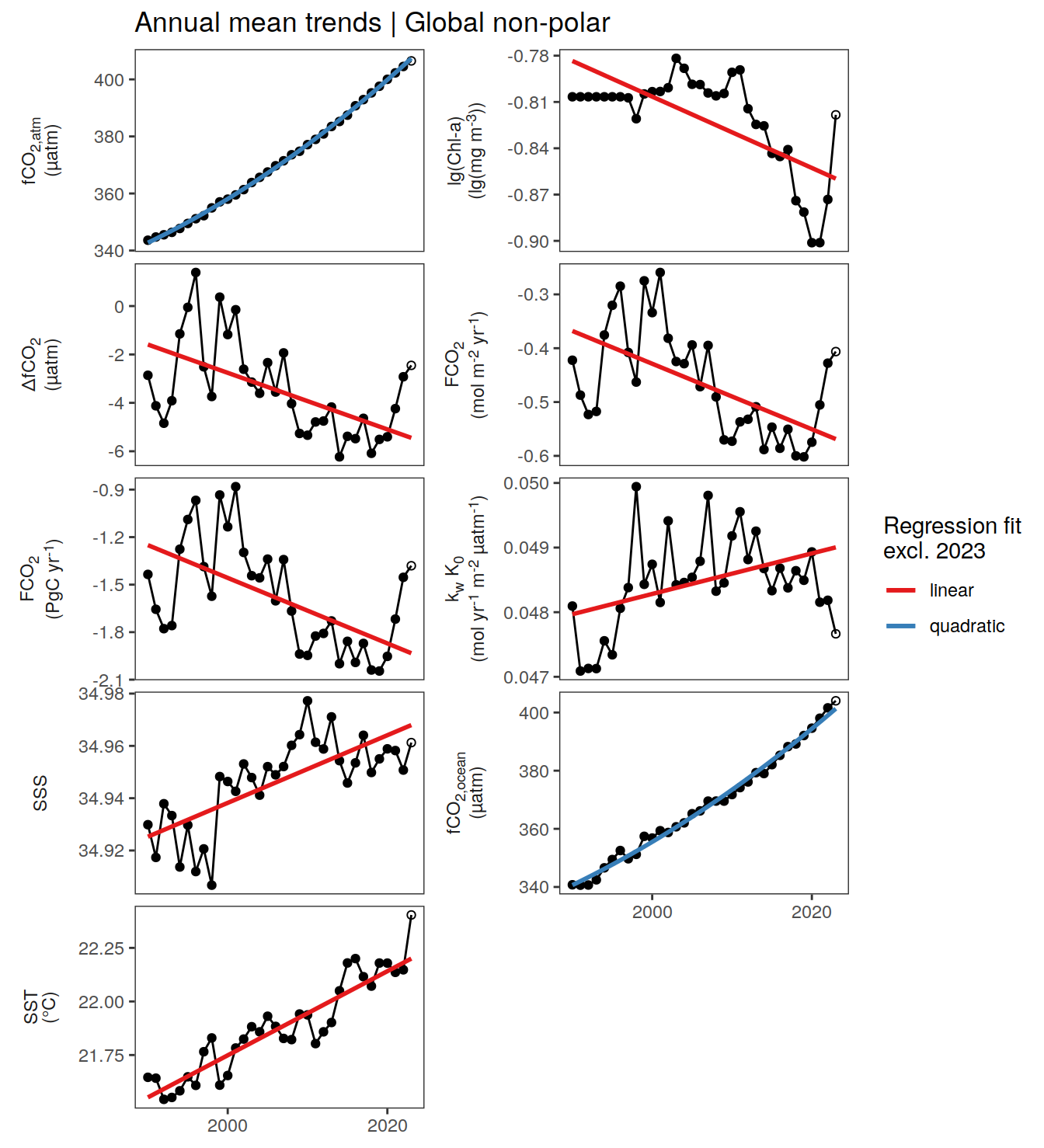
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_annual %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(year, value)) +
geom_path() +
geom_point(data = . %>% filter(year != 2023),
size = 0.2) +
geom_point(data = . %>% filter(year == 2023),
shape = 1) +
geom_smooth(data = . %>% filter(year != 2023,
!(name %in% name_quadratic_fit)),
method = "lm",
fullrange = TRUE,
aes(col = "linear"),
se = FALSE) +
geom_smooth(data = . %>% filter(year != 2023,
name %in% name_quadratic_fit),
method = "lm",
fullrange = TRUE,
formula = y ~ poly(x,2),
aes(col = "poly2"),
se = FALSE) +
scale_color_brewer(
palette = "Set1",
name = paste("Regression fit\nexcl.", 2023)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Annual mean trends | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank()
)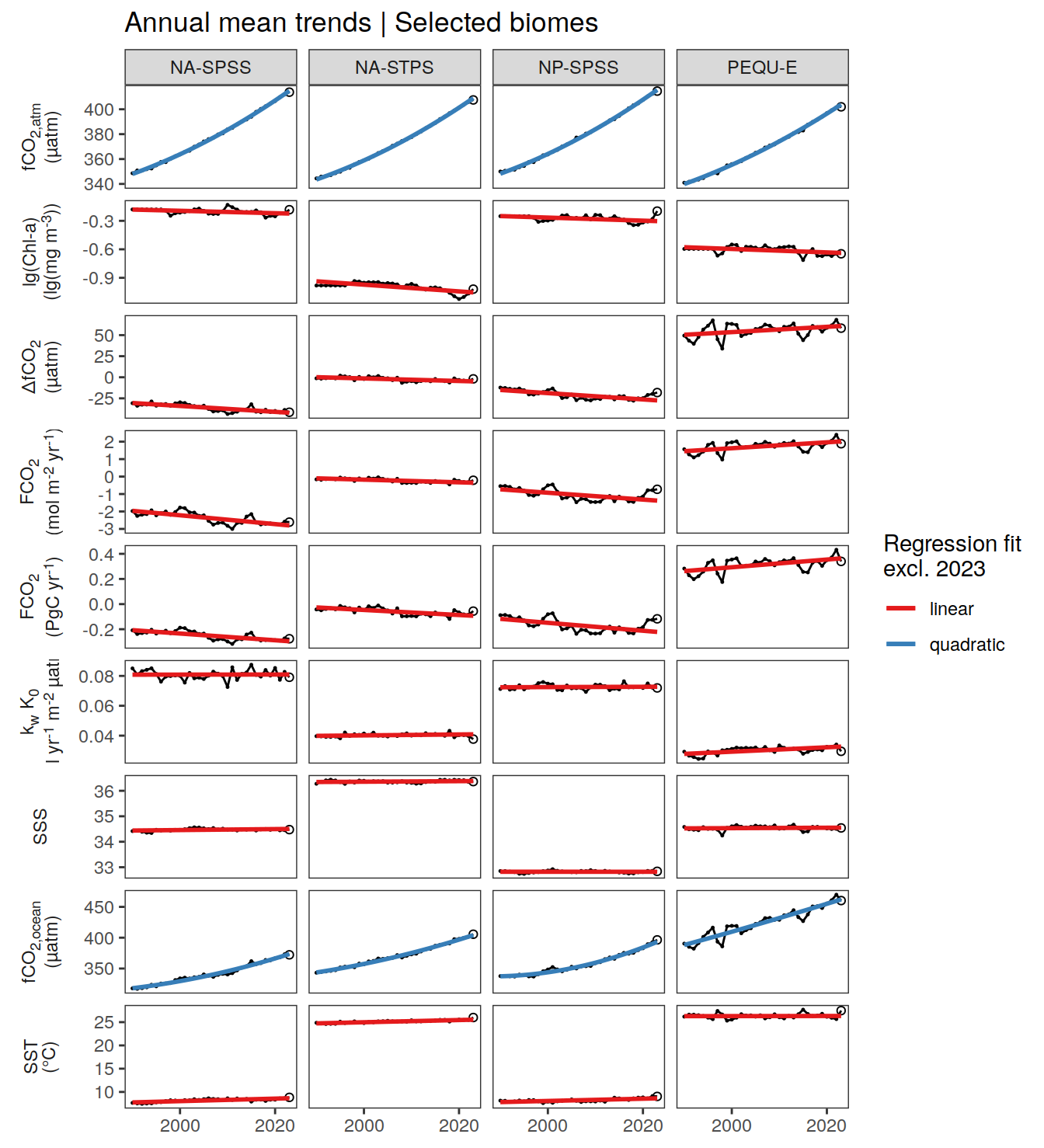
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_annual_anomaly <-
pco2_product_biome_annual %>%
anomaly_determination(biome)
pco2_product_biome_annual_anomaly %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(year, resid)) +
geom_hline(yintercept = 0) +
geom_path() +
geom_point(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(fill = year),
shape = 21) +
scale_fill_grayC() +
new_scale_fill() +
geom_point(data = . %>% filter(between(year, 2023-1, 2023)),
aes(fill = as.factor(year)),
shape = 21, size = 2) +
scale_fill_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Residual from fit to annual means (actual - predicted) | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank(),
legend.title = element_blank()
)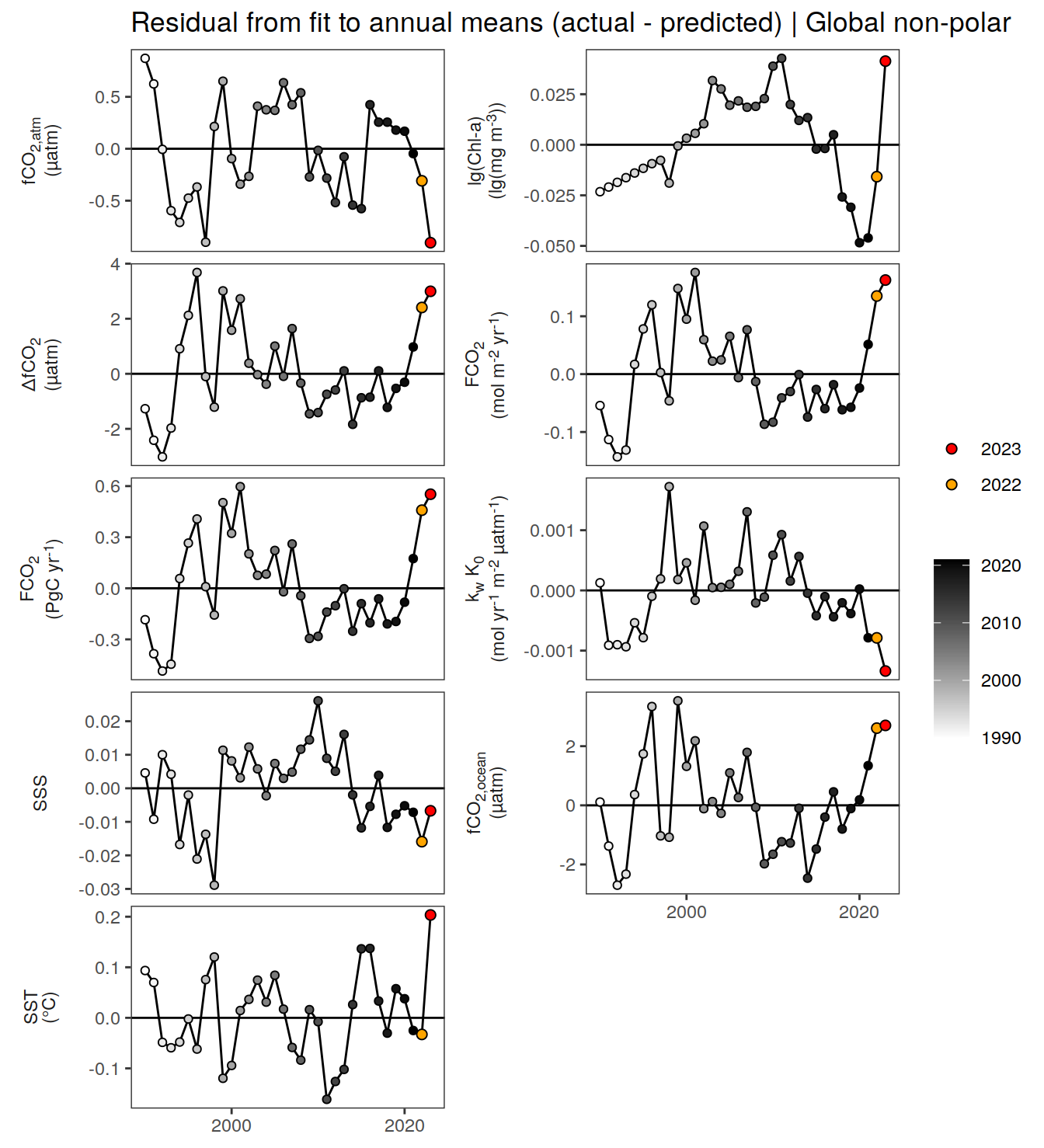
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_annual_anomaly %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(year, resid)) +
geom_hline(yintercept = 0) +
geom_path() +
geom_point(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(fill = year),
shape = 21) +
scale_fill_grayC() +
new_scale_fill() +
geom_point(data = . %>% filter(between(year, 2023-1, 2023)),
aes(fill = as.factor(year)),
shape = 21, size = 2) +
scale_fill_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1980, 2020, 20)) +
labs(title = "Residual from fit to annual means (actual - predicted) | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank(),
legend.title = element_blank()
)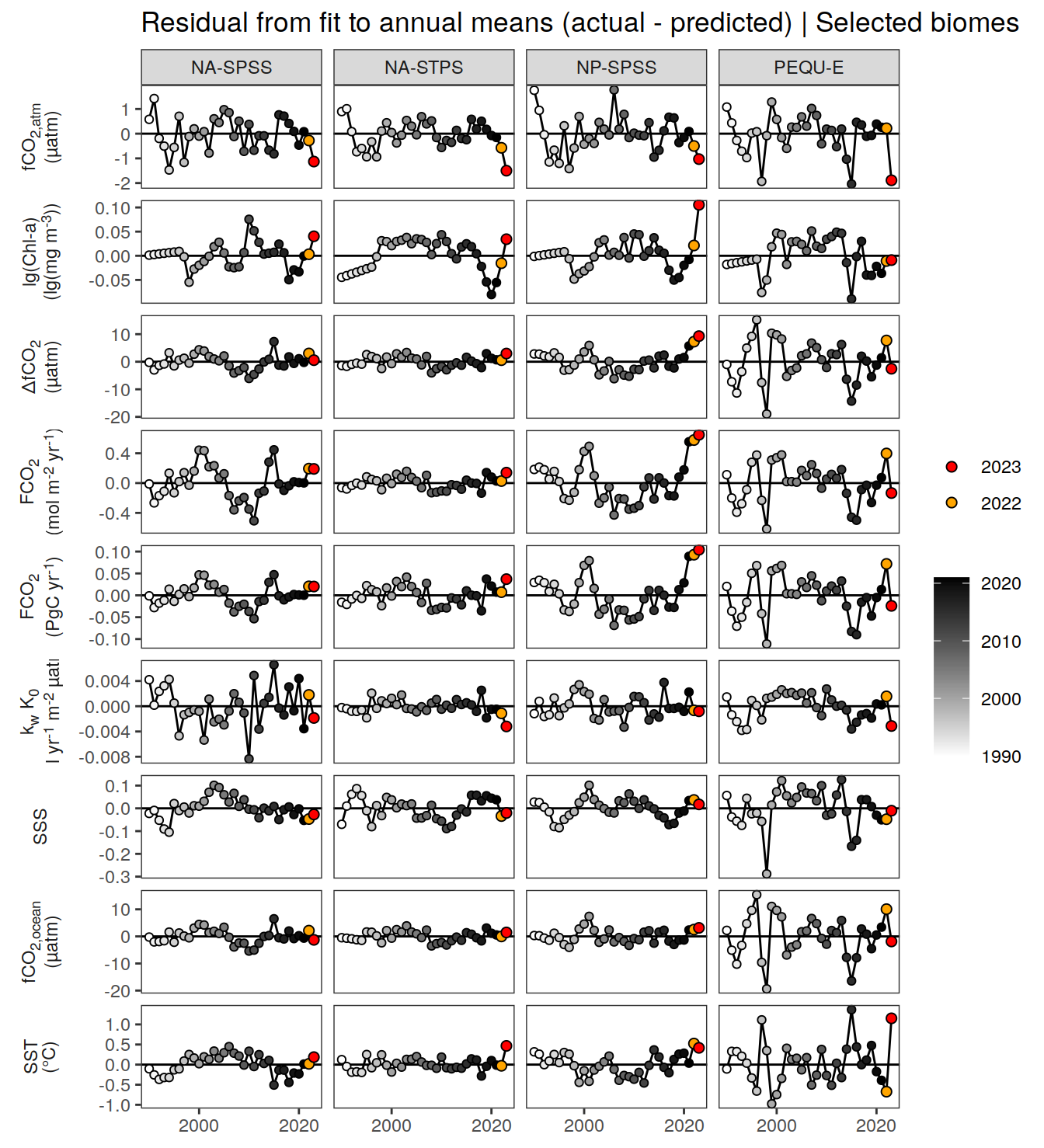
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_annual_anomaly %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_annual_anomaly.csv"
)
)pco2_product_biome_annual_detrended <-
full_join(pco2_product_biome_monthly,
pco2_product_biome_annual_anomaly %>% select(-c(value, resid))) %>%
mutate(resid = value - fit)
pco2_product_biome_annual_detrended %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted annual mean | Global non-polar") +
facet_wrap(name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)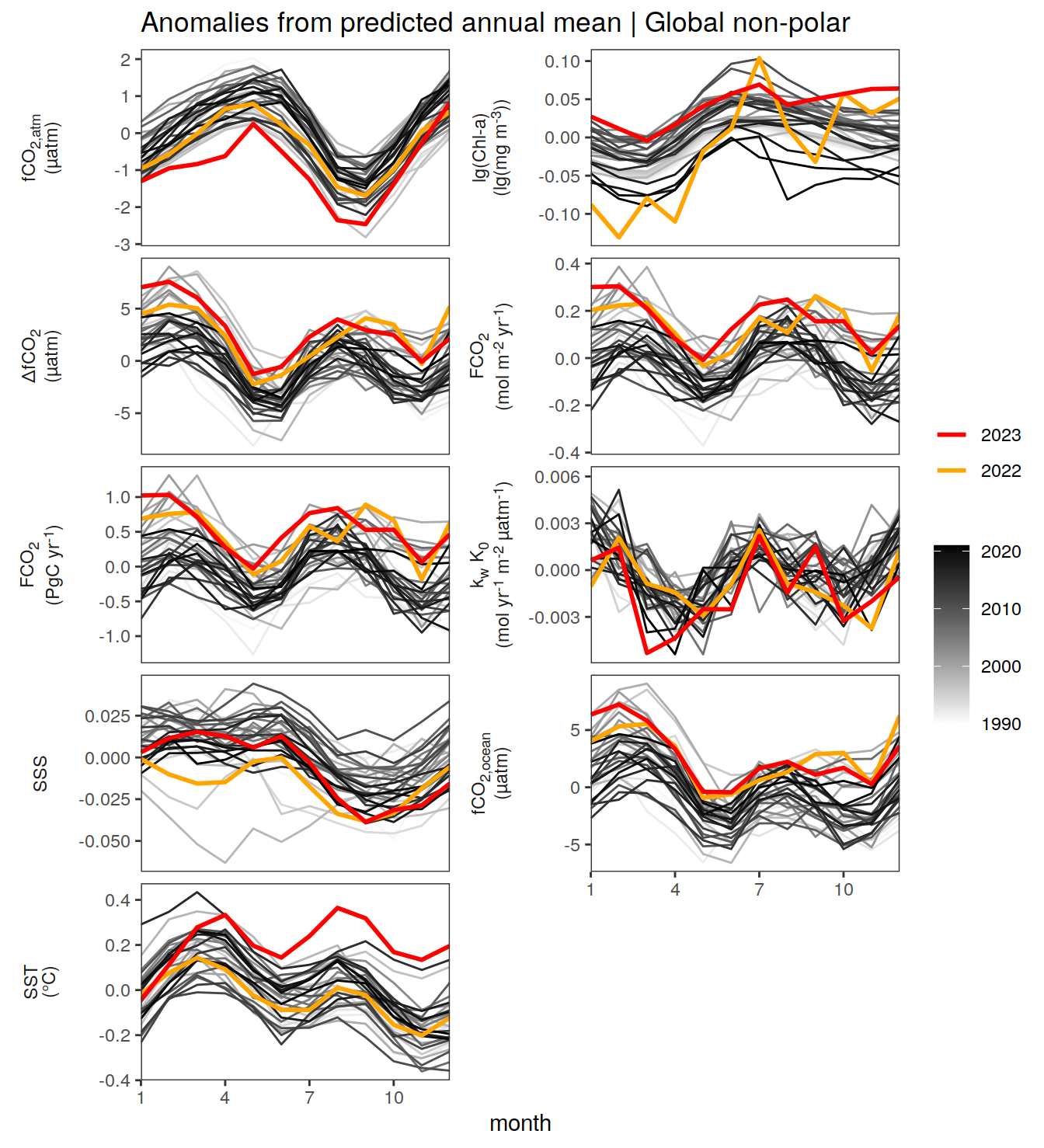
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_annual_detrended %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted annual mean | Selected biomes") +
facet_grid(name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y") +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank(),
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)
)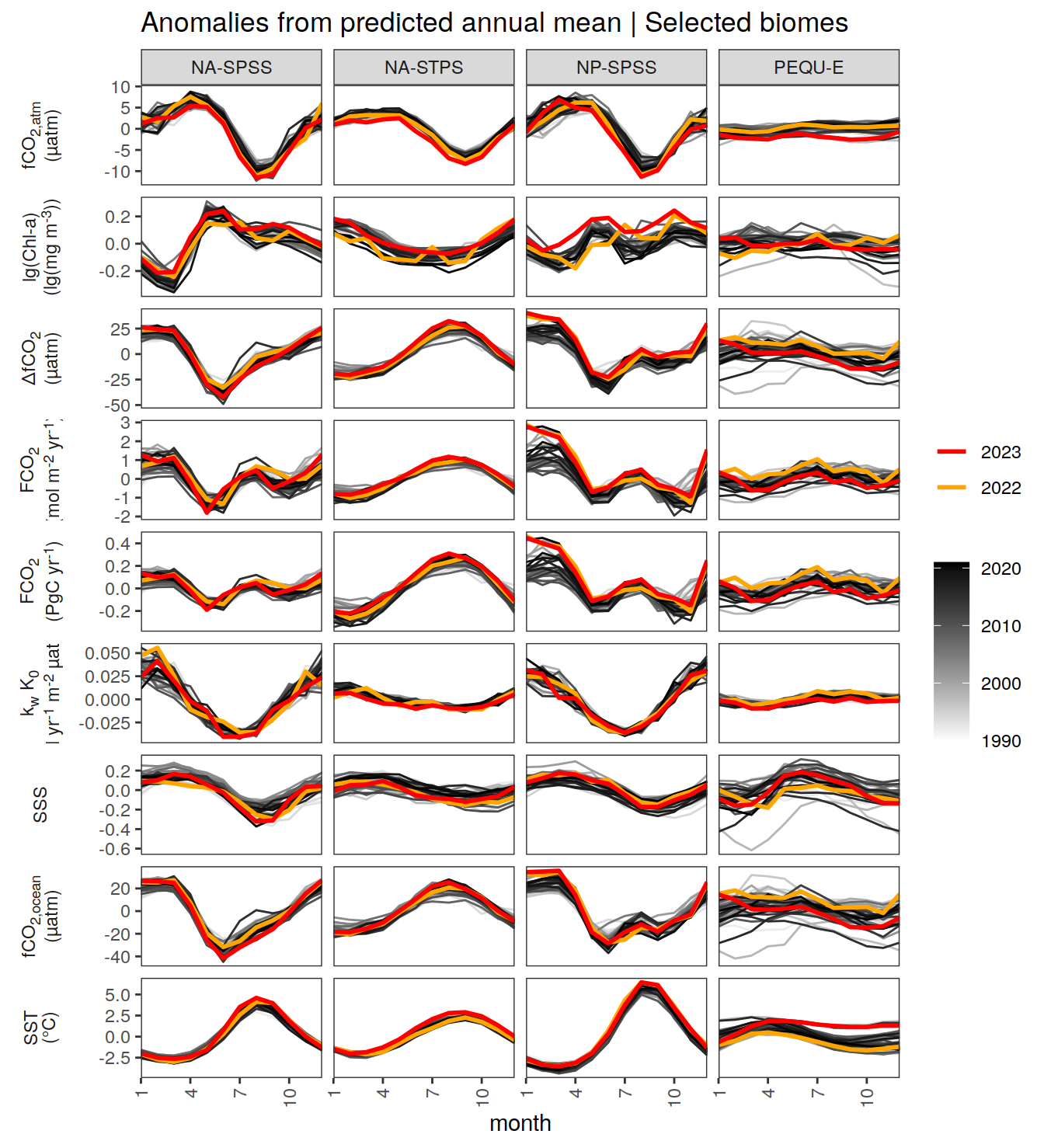
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_annual_detrended %>%
filter(biome %in% key_biomes) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(
data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1
) +
scale_color_manual(
values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = paste("Anomalies from predicted annual mean |", .x$biome)) +
facet_wrap(
name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title = element_blank(),
legend.title = element_blank()
)
)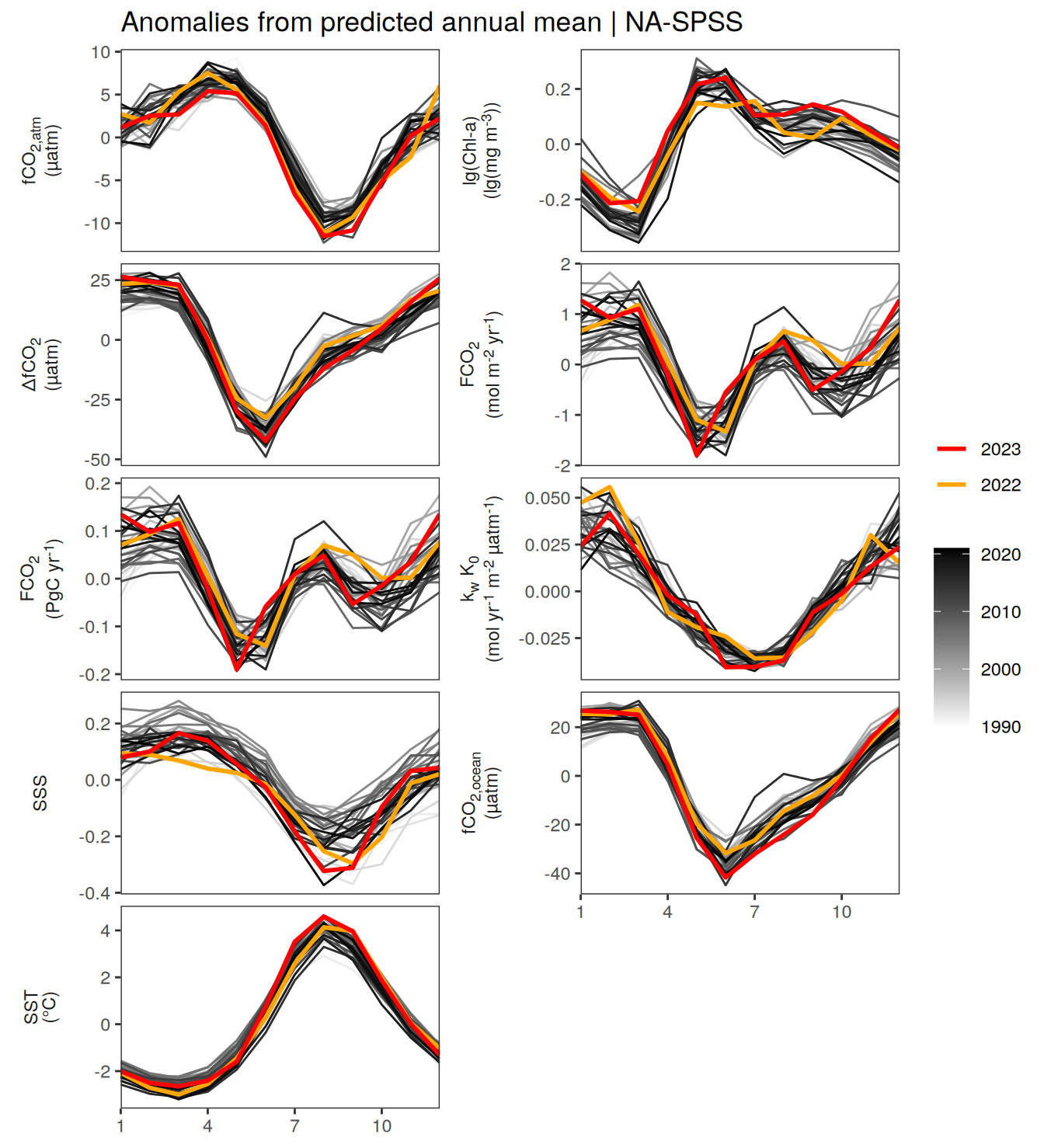
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
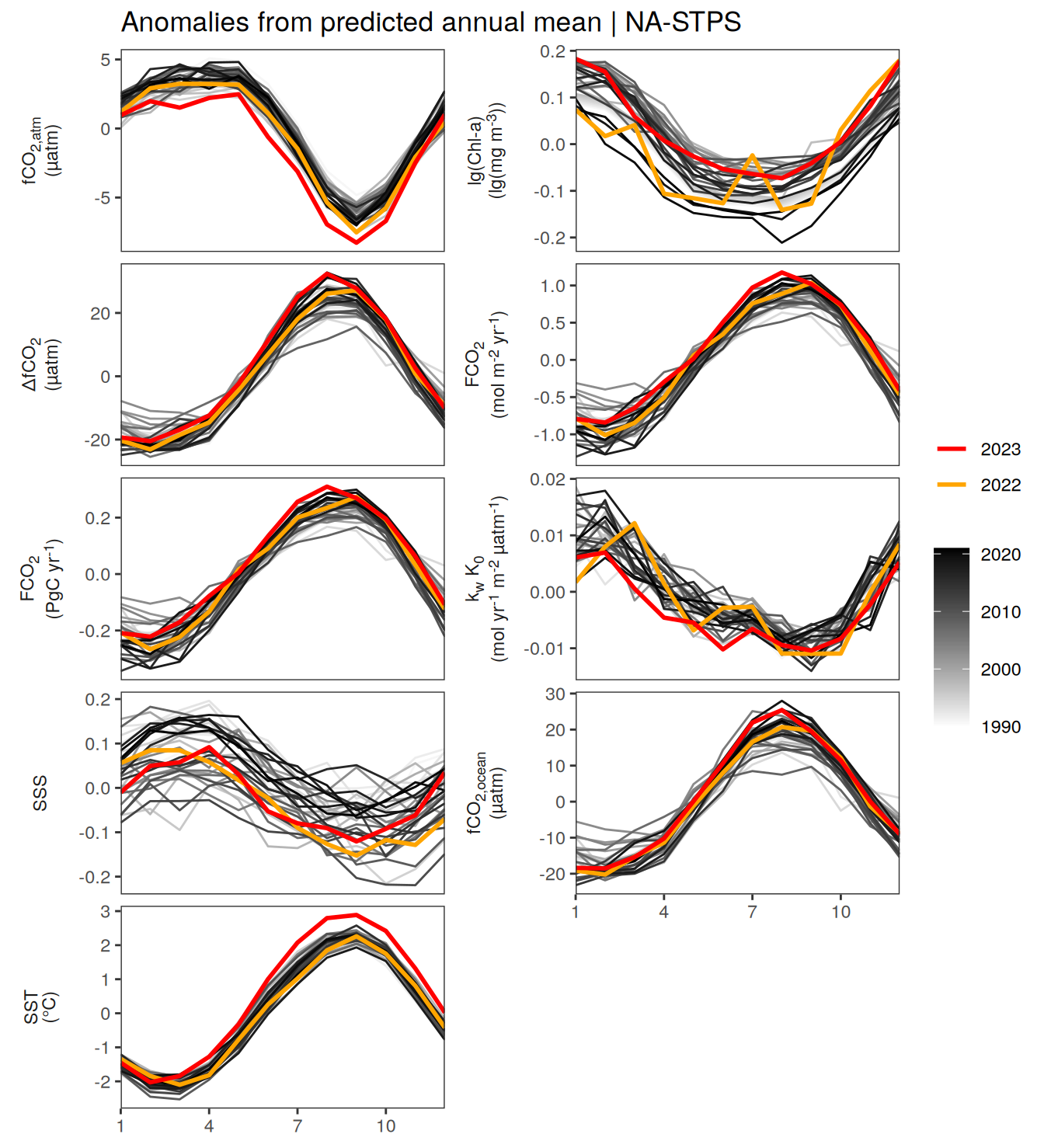
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
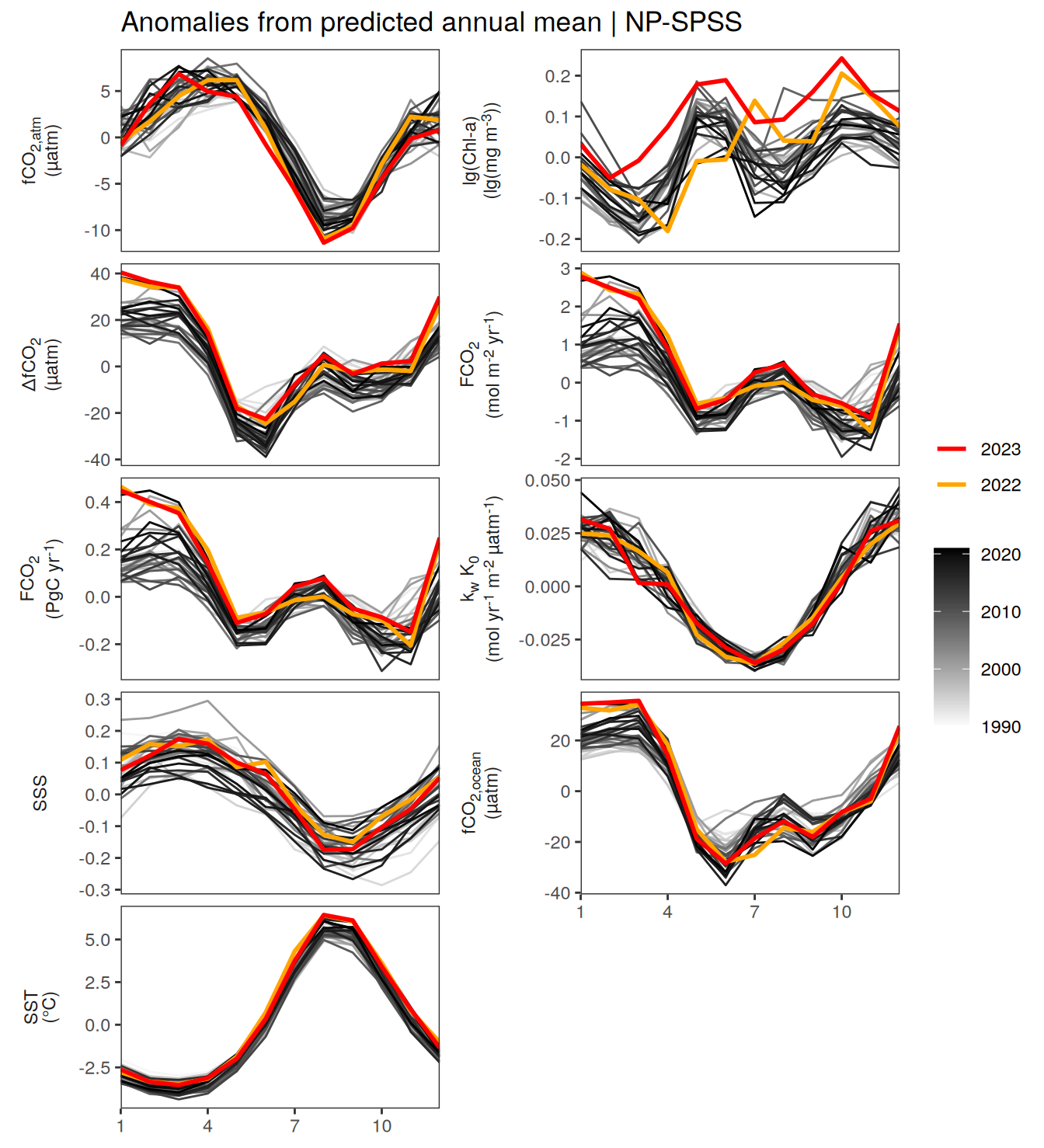
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
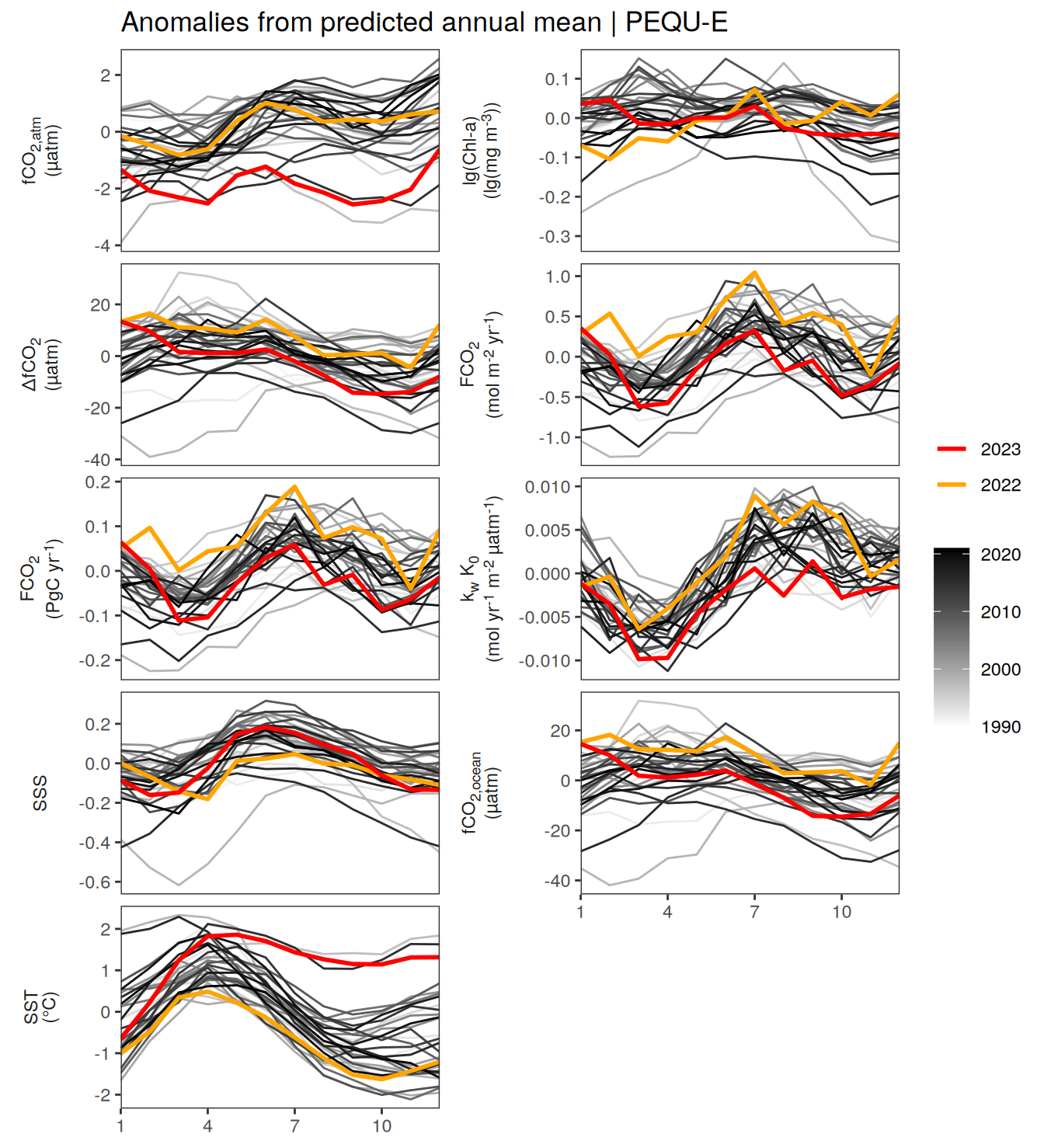
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_annual_detrended %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_annual_detrended.csv"
)
)Monthly mean trends
# pco2_product_biome_monthly %>%
# filter(biome %in% "Global non-polar") %>%
# mutate(month = as.factor(month)) %>%
# ggplot(aes(year, value, col = month)) +
# # geom_point() +
# geom_smooth(data = . %>% filter(year != 2023,
# !(name %in% name_quadratic_fit)),
# method = "lm",
# se = FALSE,
# fullrange = TRUE) +
# geom_smooth(
# data = . %>% filter(year != 2023,
# name %in% name_quadratic_fit),
# method = "lm",
# fullrange = TRUE,
# formula = y ~ x + I(x ^ 2),
# se = FALSE
# ) +
# scale_color_scico_d(palette = "romaO",
# name = paste("Regression fit\nexcl.", 2023)) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Monthly mean trends | Global non-polar") +
# facet_wrap(name ~ .,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# strip.position = "left",
# ncol = 2) +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank()
# )
#
# pco2_product_biome_monthly %>%
# filter(biome %in% key_biomes) %>%
# mutate(month = as.factor(month)) %>%
# ggplot(aes(year, value, col = month)) +
# # geom_point() +
# geom_smooth(data = . %>% filter(year != 2023,
# !(name %in% name_quadratic_fit)),
# method = "lm",
# fullrange = TRUE,
# se = FALSE) +
# geom_smooth(
# data = . %>% filter(year != 2023,
# name %in% name_quadratic_fit),
# method = "lm",
# fullrange = TRUE,
# formula = y ~ x + I(x ^ 2),
# se = FALSE
# ) +
# scale_color_scico_d(palette = "romaO",
# name = paste("Regression fit\nexcl.", 2023)) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Monthly mean trends | Selected biomes") +
# facet_grid(name ~ biome,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# switch = "y") +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank()
# )
pco2_product_biome_monthly_anomaly <-
pco2_product_biome_monthly %>%
anomaly_determination(biome, month)
# pco2_product_biome_monthly_anomaly %>%
# filter(biome %in% "Global non-polar") %>%
# group_split(month) %>%
# head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, resid)) +
# geom_hline(yintercept = 0) +
# geom_path() +
# geom_point(
# data = . %>% filter(!between(year, 2023-1, 2023)),
# aes(fill = year),
# shape = 21
# ) +
# scale_fill_grayC() +
# new_scale_fill() +
# geom_point(
# data = . %>% filter(between(year, 2023-1, 2023)),
# aes(fill = as.factor(year)),
# shape = 21,
# size = 2
# ) +
# scale_fill_manual(
# values = c("orange", "red"),
# guide = guide_legend(reverse = TRUE,
# order = 1)
# ) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Residual from fit to monthly means (actual - predicted) | Global non-polar",
# subtitle = paste("Month:", .x$month)) +
# facet_wrap(
# name ~ .,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# strip.position = "left",
# ncol = 2
# ) +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank(),
# legend.title = element_blank()
# )
# )
#
# pco2_product_biome_monthly_anomaly %>%
# filter(biome %in% key_biomes) %>%
# group_split(month) %>%
# head(1) %>%
# map(
# ~ ggplot(data = .x,
# aes(year, resid)) +
# geom_hline(yintercept = 0) +
# geom_path() +
# geom_point(
# data = . %>% filter(!between(year, 2023-1, 2023)),
# aes(fill = year),
# shape = 21
# ) +
# scale_fill_grayC() +
# new_scale_fill() +
# geom_point(
# data = . %>% filter(between(year, 2023-1, 2023)),
# aes(fill = as.factor(year)),
# shape = 21,
# size = 2
# ) +
# scale_fill_manual(
# values = c("orange", "red"),
# guide = guide_legend(reverse = TRUE,
# order = 1)
# ) +
# scale_x_continuous(breaks = seq(1980, 2020, 20)) +
# labs(title = "Residual from fit to monthly means (actual - predicted) | Selected biomes",
# subtitle = paste("Month:", .x$month)) +
# facet_grid(
# name ~ biome,
# scales = "free_y",
# labeller = labeller(name = x_axis_labels),
# switch = "y"
# ) +
# theme(
# strip.text.y.left = element_markdown(),
# strip.placement = "outside",
# strip.background.y = element_blank(),
# axis.title = element_blank(),
# legend.title = element_blank()
# )
# )pco2_product_biome_monthly_anomaly %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_monthly_anomaly.csv"
)
)Global non-polar
pco2_product_biome_monthly_detrended <-
full_join(pco2_product_biome_monthly,
pco2_product_biome_monthly_anomaly %>% select(-c(value, resid))) %>%
mutate(resid = value - fit)
pco2_product_biome_monthly_detrended %>%
filter(biome %in% "Global non-polar") %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted monthly mean | Global non-polar") +
facet_wrap(
name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)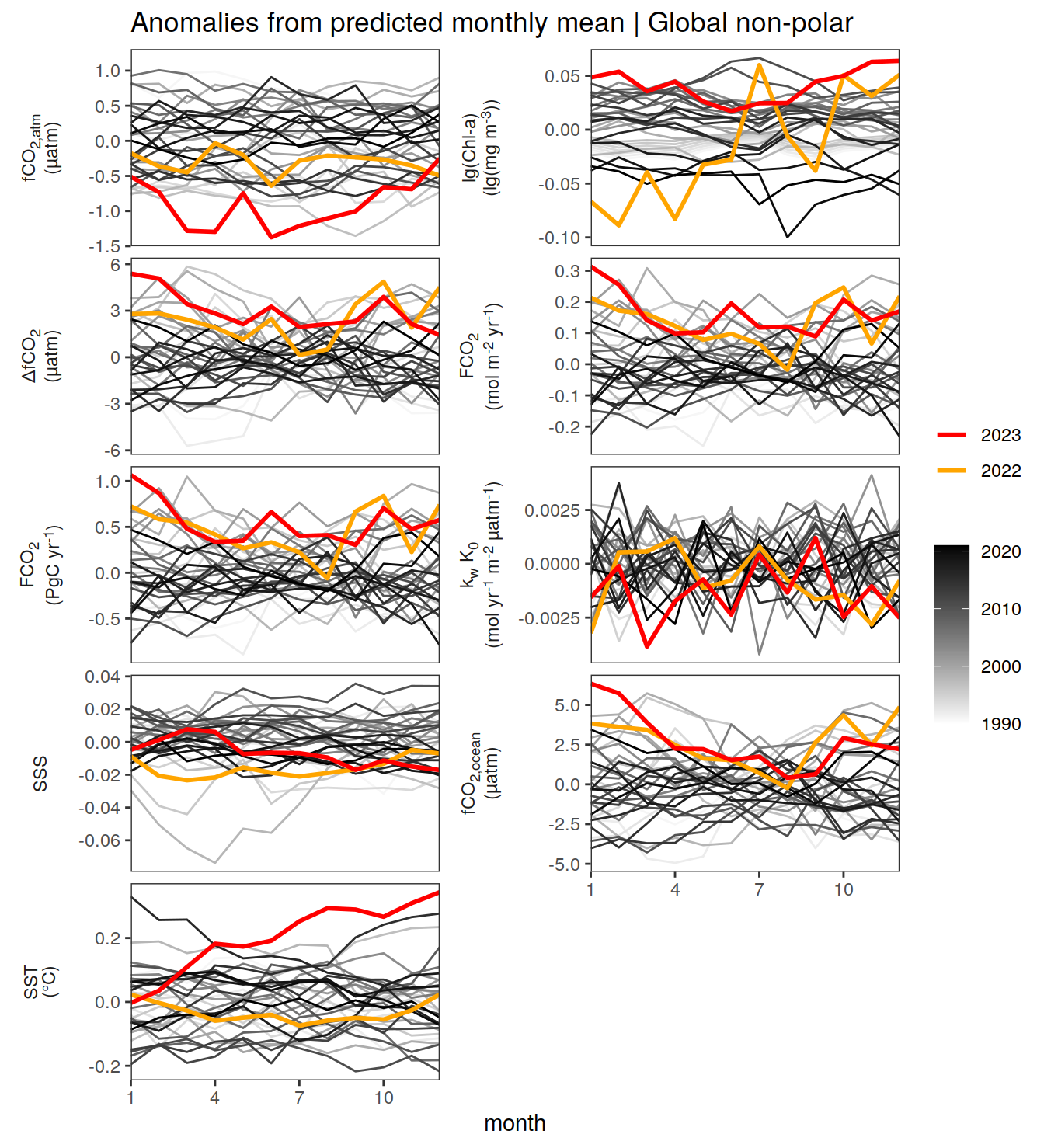
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_monthly_detrended %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted monthly mean | Selected biomes") +
facet_grid(
name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)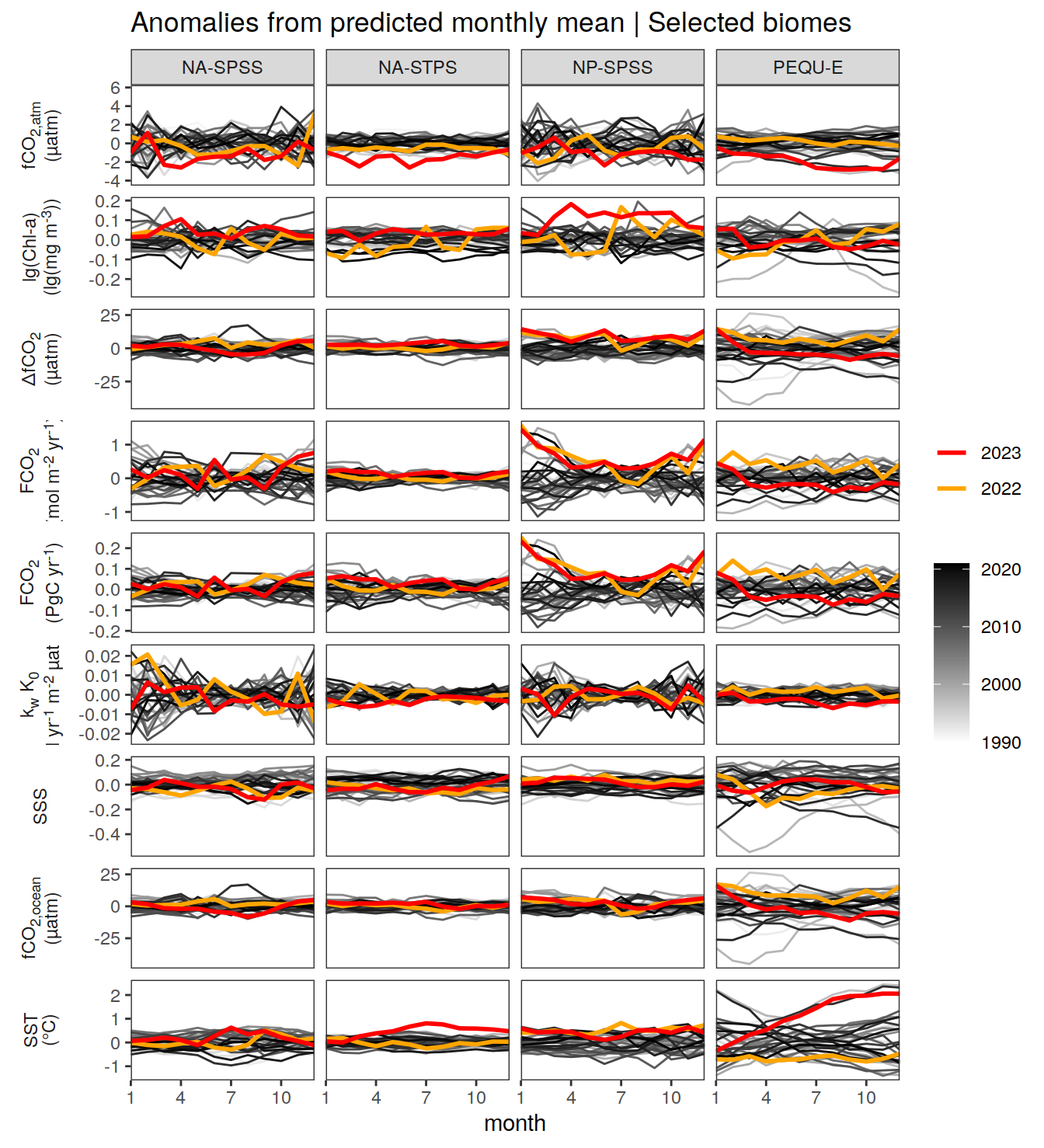
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
Key biomes
pco2_product_biome_monthly_detrended %>%
filter(biome %in% key_biomes) %>%
ggplot(aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1) +
scale_color_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = "Anomalies from predicted monthly mean | Selected biomes") +
facet_grid(
name ~ biome,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
switch = "y"
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)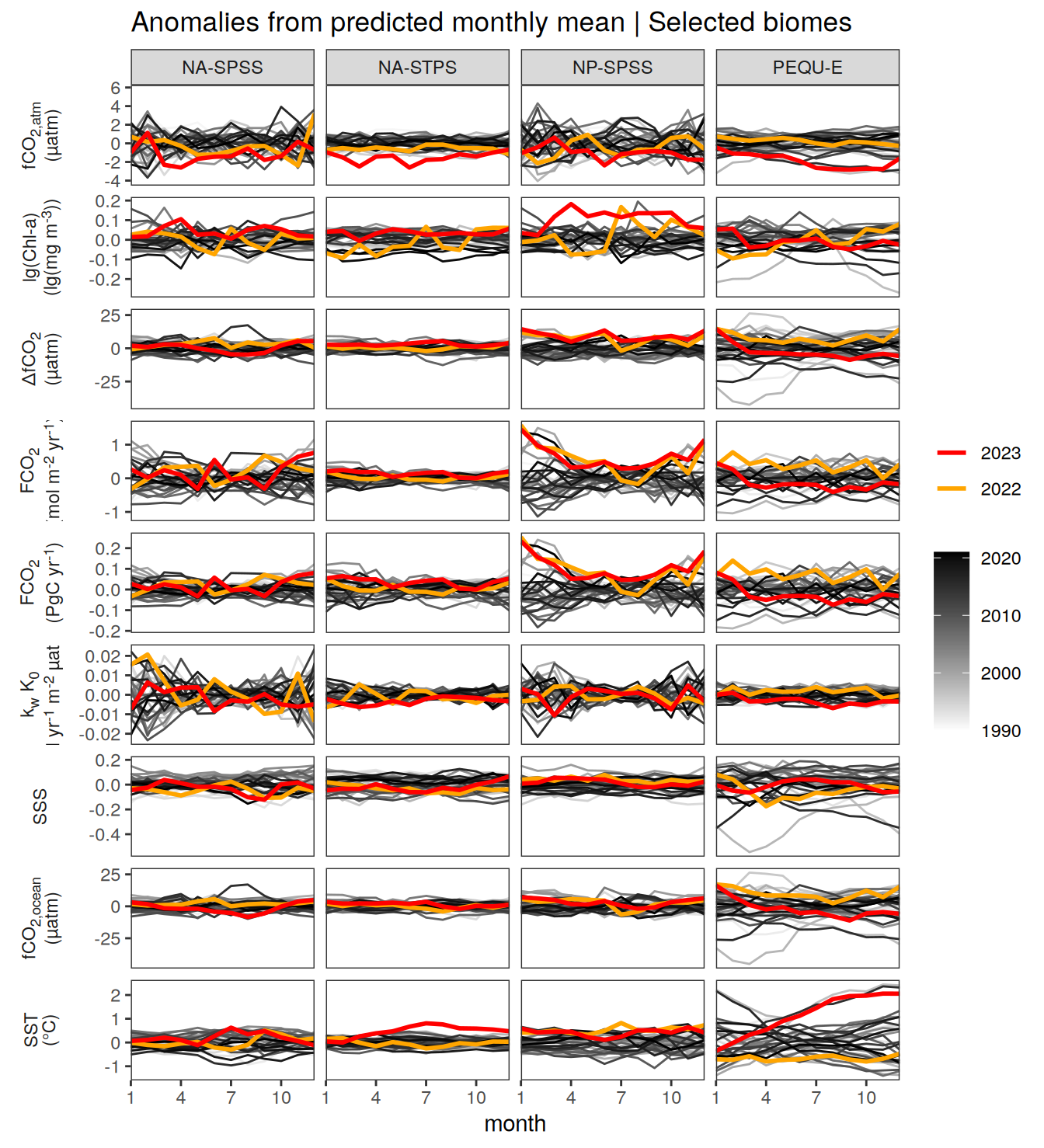
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_monthly_detrended %>%
filter(biome %in% key_biomes) %>%
group_split(biome) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(month, resid, group = as.factor(year))) +
geom_path(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(col = year)) +
scale_color_grayC() +
new_scale_color() +
geom_path(
data = . %>% filter(between(year, 2023-1, 2023)),
aes(col = as.factor(year)),
linewidth = 1
) +
scale_color_manual(
values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)
) +
scale_x_continuous(breaks = seq(1, 12, 3), expand = c(0, 0)) +
labs(title = paste("Anomalies from predicted monthly mean |", .x$biome)) +
facet_wrap(
name ~ .,
scales = "free_y",
labeller = labeller(name = x_axis_labels),
strip.position = "left",
ncol = 2
) +
theme(
strip.text.y.left = element_markdown(),
strip.placement = "outside",
strip.background.y = element_blank(),
axis.title.y = element_blank(),
legend.title = element_blank()
)
)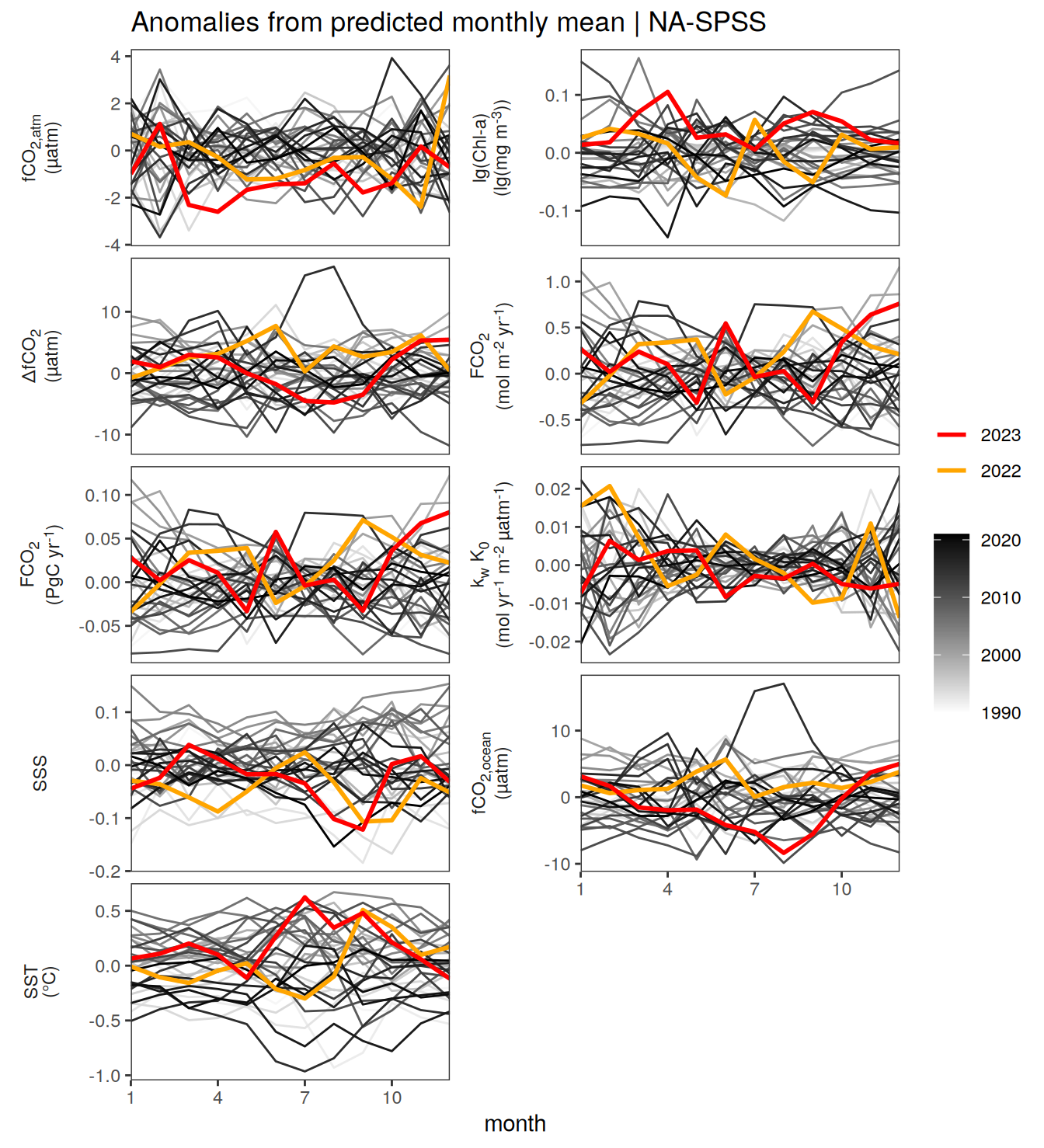
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
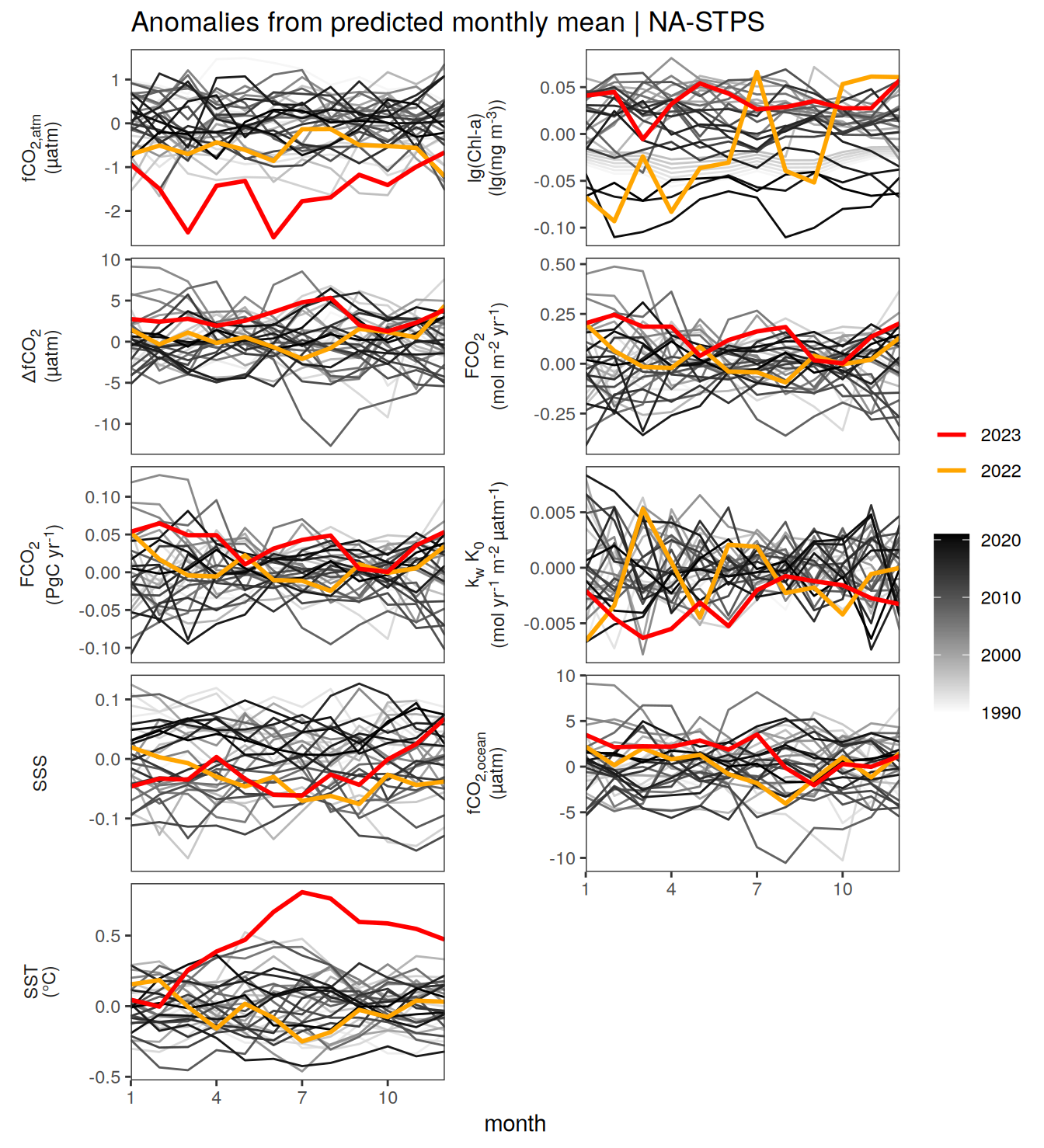
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
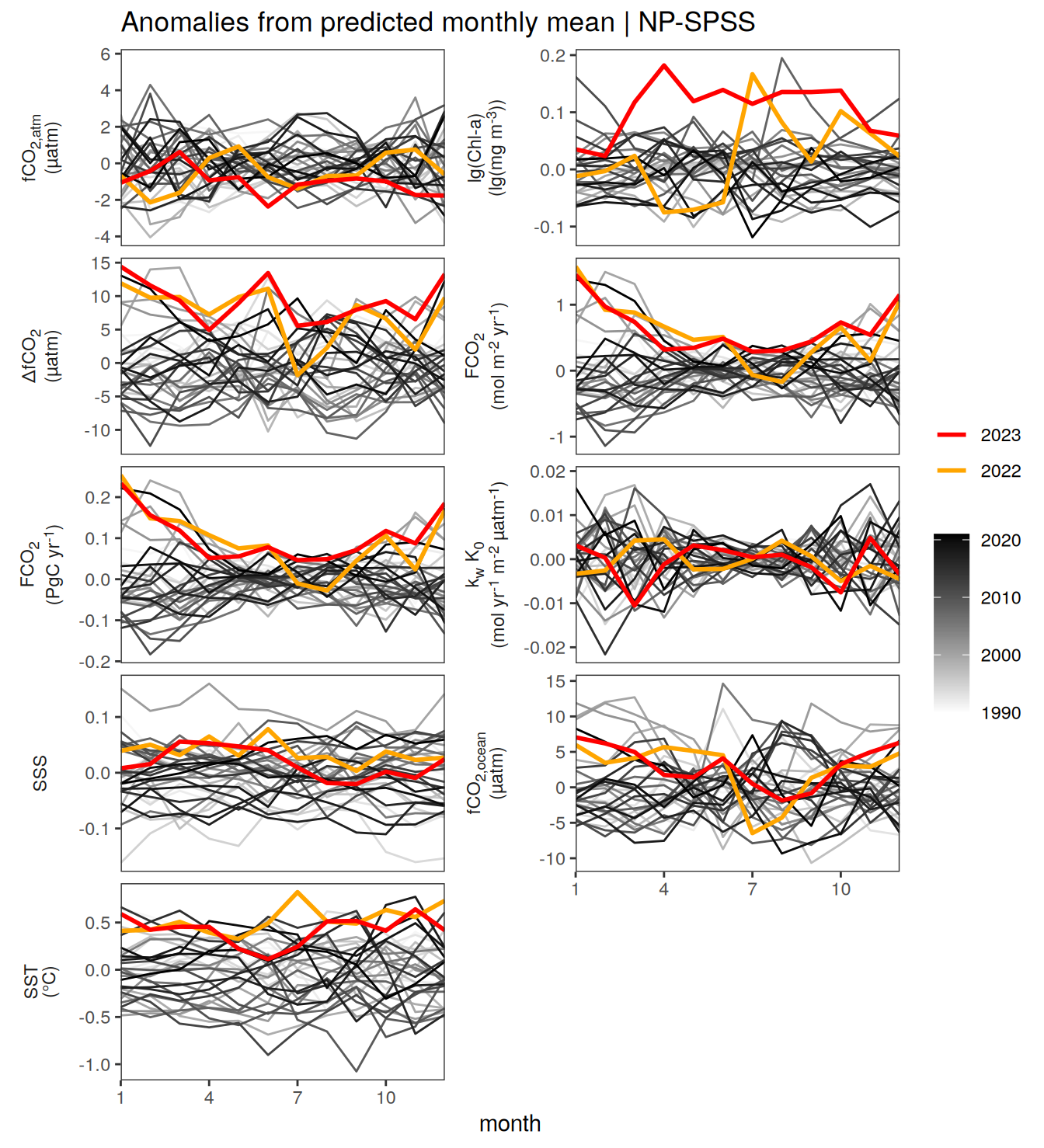
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
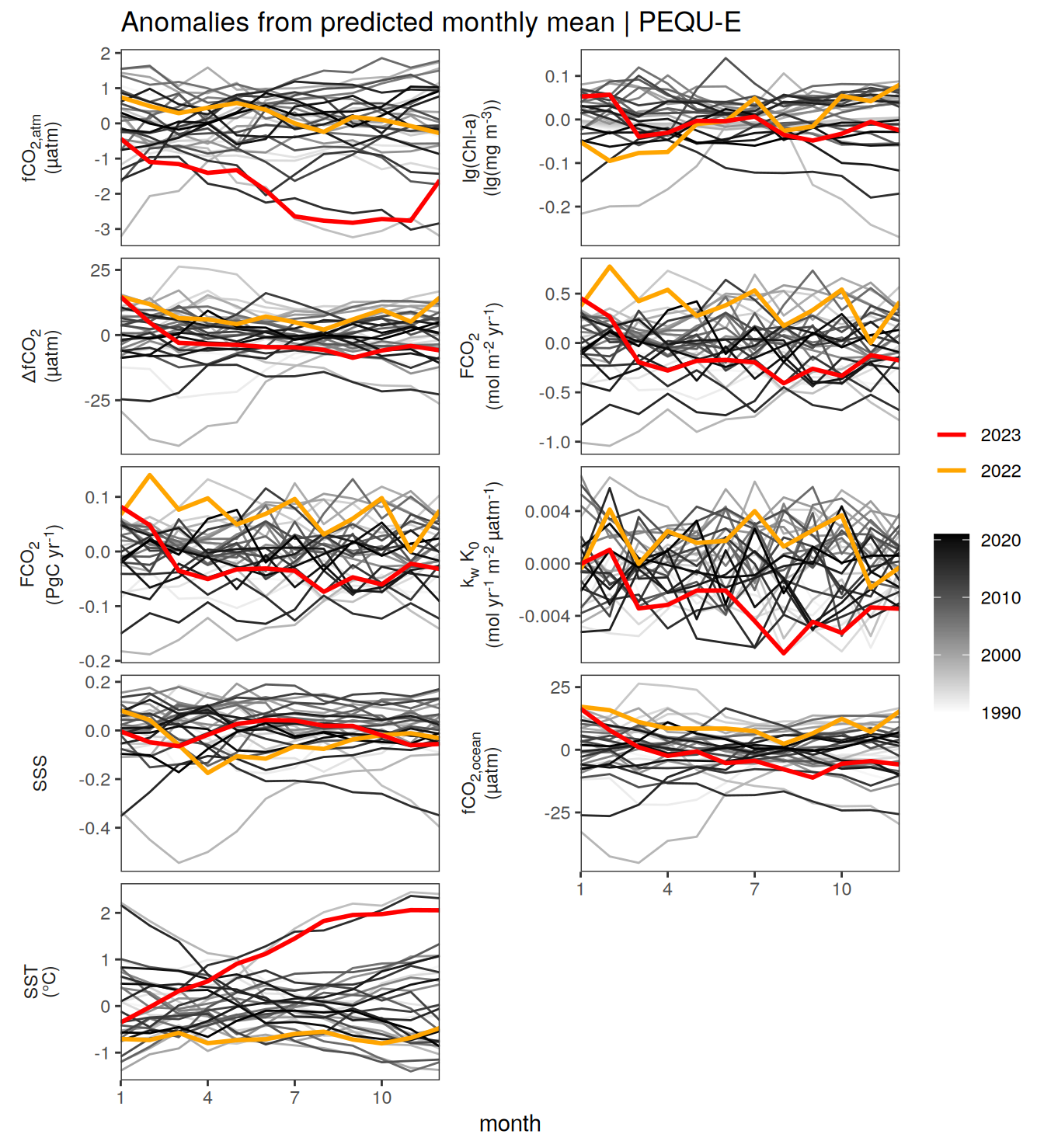
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_monthly_detrended %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_monthly_detrended.csv"
)
)2023 anomaly correlation
The following plots aim to unravel the correlation between regionally integrated monthly flux anomalies and the corresponding anomalies of the means/integrals of each other variable.
Anomalies are first presented are first presented in absolute units. Due to the different flux magnitudes, we need to plot integrated fluxes separately for each region. Secondly, we normalize the monthly anomalies to the spread (expressed as standard deviation) of the residuals from the fit.
Annual anomalies
Absolute
pco2_product_biome_annual_anomaly %>%
filter(biome == "Global non-polar") %>%
select(-c(value, fit)) %>%
pivot_wider(values_from = resid) %>%
pivot_longer(-c(year, biome, fgco2_int)) %>%
ggplot(aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(data = . %>% filter(!between(year, 2023-1, 2023)),
aes(fill = year),
shape = 21) +
geom_smooth(
data = . %>% filter(!between(year, 2023-1, 2023)),
method = "lm",
se = FALSE,
fullrange = TRUE,
aes(col = paste("Regression fit\nexcl.", 2023))
) +
scale_color_grey() +
scale_fill_grayC()+
new_scale_fill() +
geom_point(data = . %>% filter(between(year, 2023-1, 2023)),
aes(fill = as.factor(year)),
shape = 21, size = 2) +
scale_fill_manual(values = c("orange", "red"),
guide = guide_legend(reverse = TRUE,
order = 1)) +
labs(title = "Global non-polar integrated fluxes",
y = labels_breaks("fgco2_int")$i_legend_title) +
facet_wrap(
~ name,
scales = "free_x",
labeller = labeller(name = x_axis_labels),
strip.position = "bottom",
ncol = 2
) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.y = element_markdown(),
axis.title.x = element_blank(),
legend.title = element_blank()
)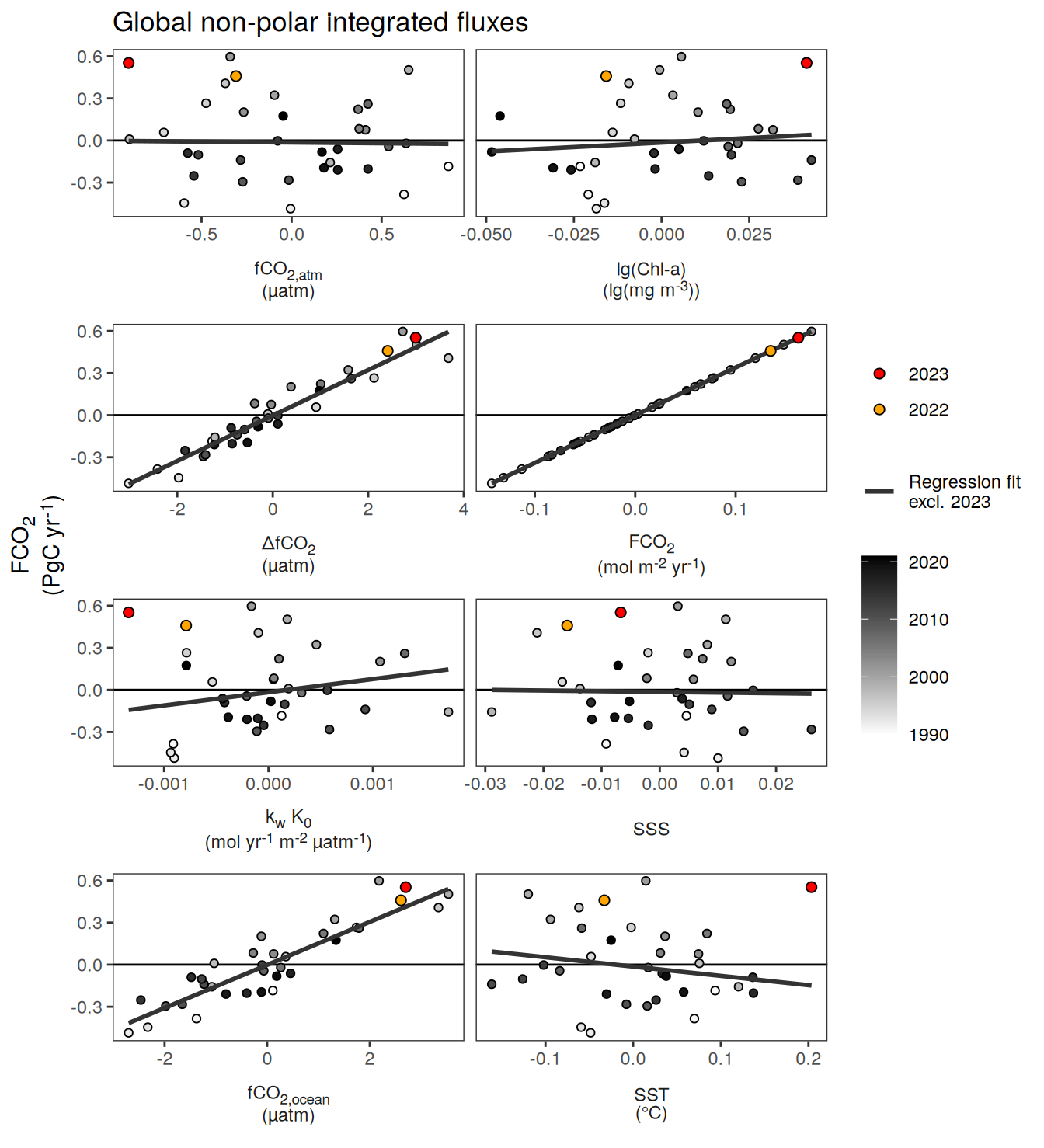
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
Monthly anomalies
Absolute
pco2_product_biome_monthly_detrended_anomaly <-
pco2_product_biome_monthly_detrended %>%
select(year, month, biome, name, resid) %>%
pivot_wider(names_from = name,
values_from = resid)
pco2_product_biome_monthly_detrended_anomaly %>%
filter(biome == "Global non-polar") %>%
pivot_longer(-c(year, month, biome, fgco2_int)) %>%
ggplot(aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3) +
scale_color_scico_d(palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)) +
scale_fill_scico_d(palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)) +
labs(title = "Global non-polar integrated fluxes",
y = labels_breaks("fgco2_int")$i_legend_title) +
facet_wrap(
~ name,
scales = "free_x",
labeller = labeller(name = x_axis_labels),
strip.position = "bottom",
ncol = 2
) +
theme(
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
axis.title.y = element_markdown(),
axis.title.x = element_blank()
)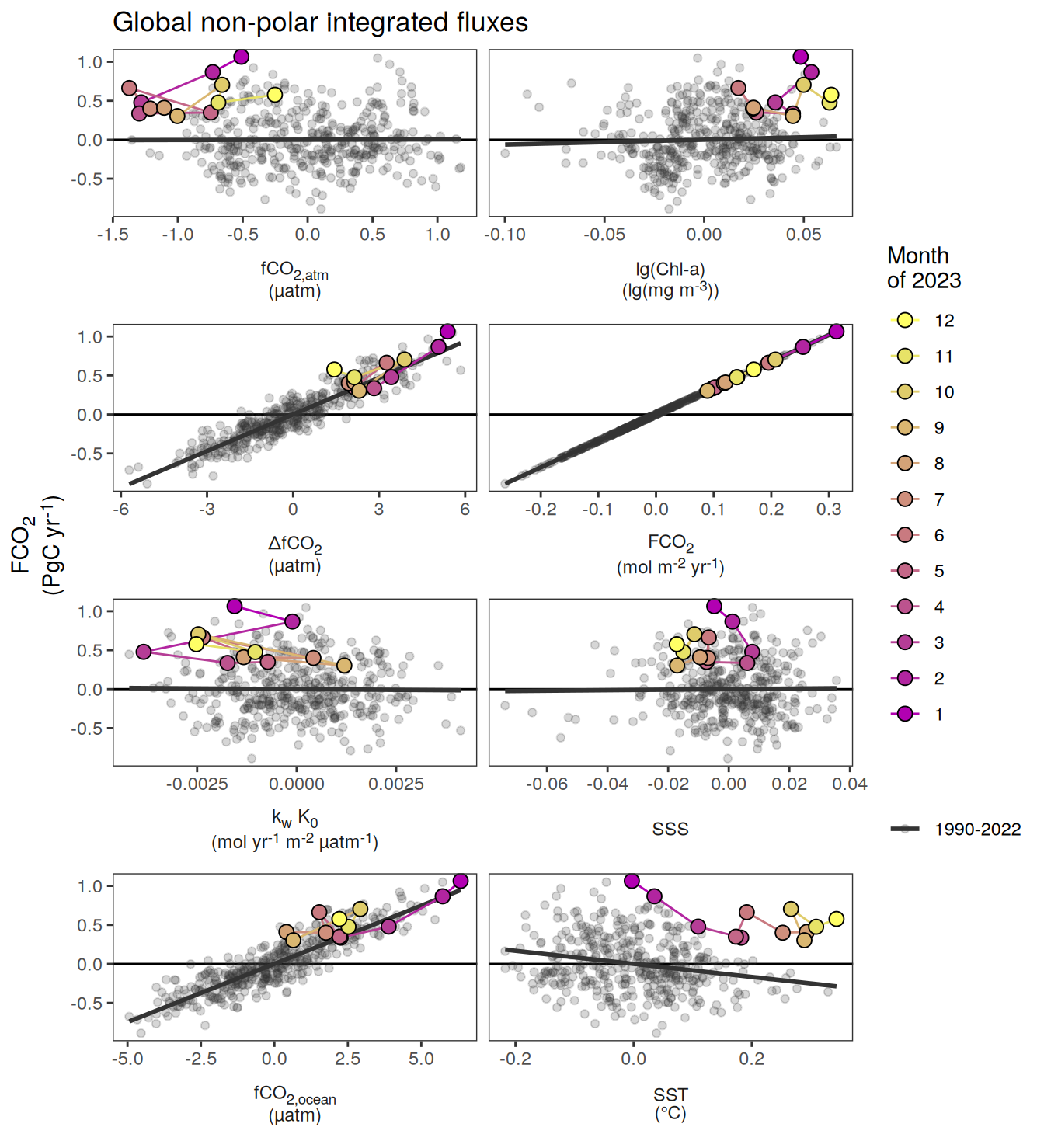
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| e1e0ccb | jens-daniel-mueller | 2024-05-27 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| be285dc | jens-daniel-mueller | 2024-05-21 |
| 51df30d | jens-daniel-mueller | 2024-05-15 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| ce4e2a6 | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
pco2_product_biome_monthly_detrended_anomaly %>%
filter(!(biome %in% c(key_biomes, "Global non-polar"))) %>%
pivot_longer(-c(year, month, biome, fgco2_int)) %>%
filter(name %in% c("temperature", "chl", "dfco2", "kw_sol")) %>%
group_split(name) %>%
head(1) %>%
map(
~ ggplot(data = .x,
aes(value, fgco2_int)) +
geom_hline(yintercept = 0) +
geom_point(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2
) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(
data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3
) +
scale_color_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
scale_fill_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
facet_wrap( ~ biome, ncol = 3, scales = "free") +
labs(
title = "Biome integrated fluxes",
y = labels_breaks("fgco2_int")$i_legend_title,
x = labels_breaks(.x %>% distinct(name))$i_legend_title
) +
theme(axis.title.x = element_markdown(),
axis.title.y = element_markdown())
)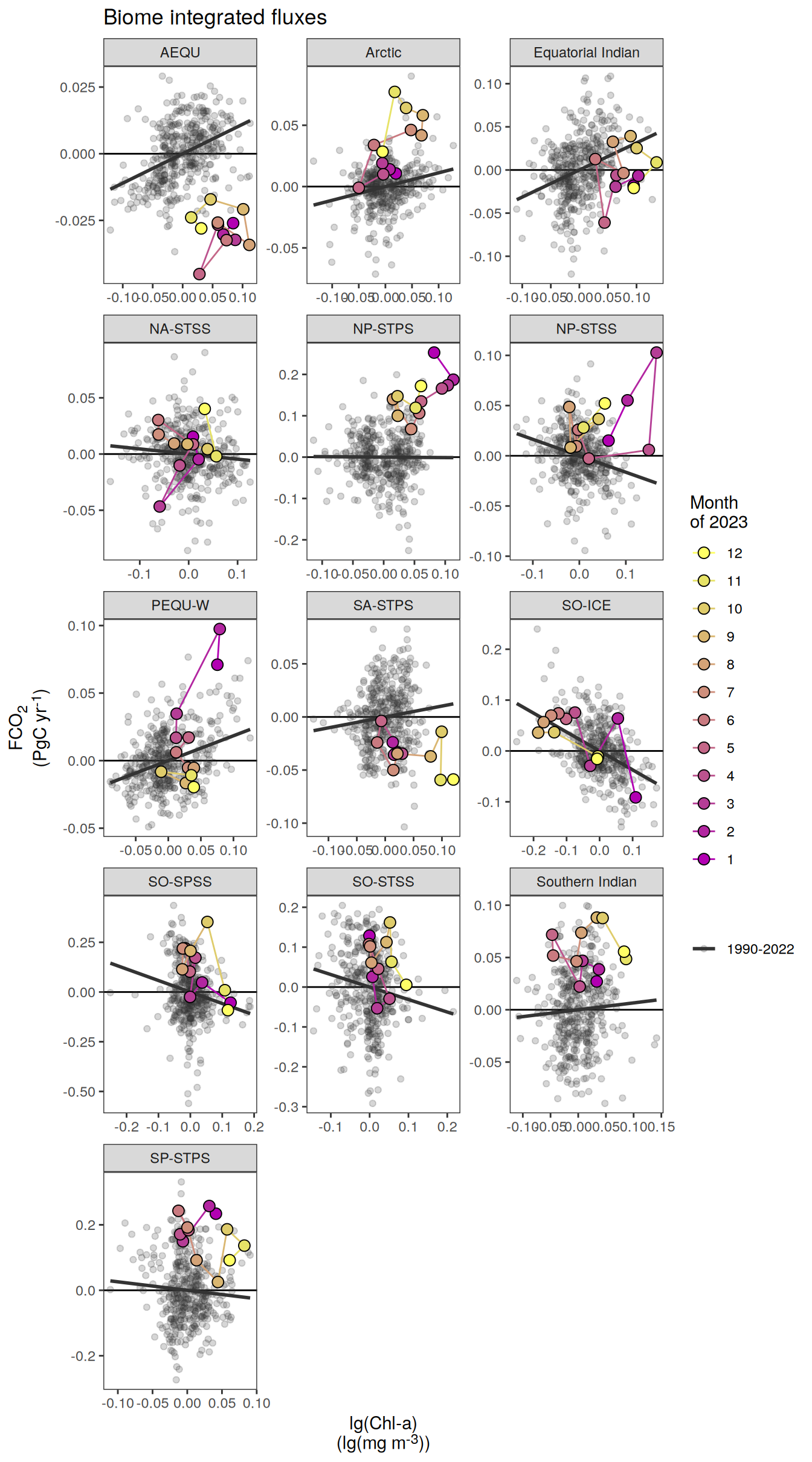
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| 1ea301d | jens-daniel-mueller | 2024-05-06 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 231f7cd | jens-daniel-mueller | 2024-04-17 |
| 741ee62 | jens-daniel-mueller | 2024-04-17 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
Relative to spread
pco2_product_biome_monthly_detrended_anomaly_spread <-
pco2_product_biome_monthly_detrended_anomaly %>%
pivot_longer(-c(month, biome, year)) %>%
filter(year != 2023) %>%
group_by(month, biome, name) %>%
summarise(spread = sd(value, na.rm = TRUE)) %>%
ungroup()
pco2_product_biome_monthly_detrended_anomaly_relative <-
full_join(
pco2_product_biome_monthly_detrended_anomaly_spread,
pco2_product_biome_monthly_detrended_anomaly %>%
pivot_longer(-c(month, biome, year))
)
pco2_product_biome_monthly_detrended_anomaly_relative <-
pco2_product_biome_monthly_detrended_anomaly_relative %>%
mutate(value = value / spread) %>%
select(-spread) %>%
pivot_wider() %>%
pivot_longer(-c(month, biome, year, fgco2_int))
pco2_product_biome_monthly_detrended_anomaly_relative %>%
filter(name %in% c("temperature", "chl", "dfco2", "kw_sol")) %>%
group_split(name) %>%
head(1) %>%
map(
~ ggplot(data = .x,
aes(value, fgco2_int)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 0) +
geom_point(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
alpha = 0.2
) +
geom_smooth(
data = . %>% filter(year != 2023),
aes(col = paste(min(year), max(year), sep = "-")),
method = "lm",
se = FALSE,
fullrange = TRUE
) +
scale_color_grey(name = "") +
new_scale_color() +
geom_path(data = . %>% filter(year == 2023),
aes(col = as.factor(month), group = 1)) +
geom_point(
data = . %>% filter(year == 2023),
aes(fill = as.factor(month)),
shape = 21,
size = 3
) +
scale_color_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
scale_fill_scico_d(
palette = "buda",
guide = guide_legend(reverse = TRUE,
order = 1),
name = paste("Month\nof", 2023)
) +
facet_wrap( ~ biome, ncol = 3) +
coord_fixed() +
labs(
title = "Biome integrated fluxes normalized to spread",
y = str_split_i(labels_breaks("fgco2_int")$i_legend_title, "<br>", i = 1),
x = str_split_i(labels_breaks(.x %>% distinct(name))$i_legend_title, "<br>", i = 1)
) +
theme(axis.title.x = element_markdown(),
axis.title.y = element_markdown())
)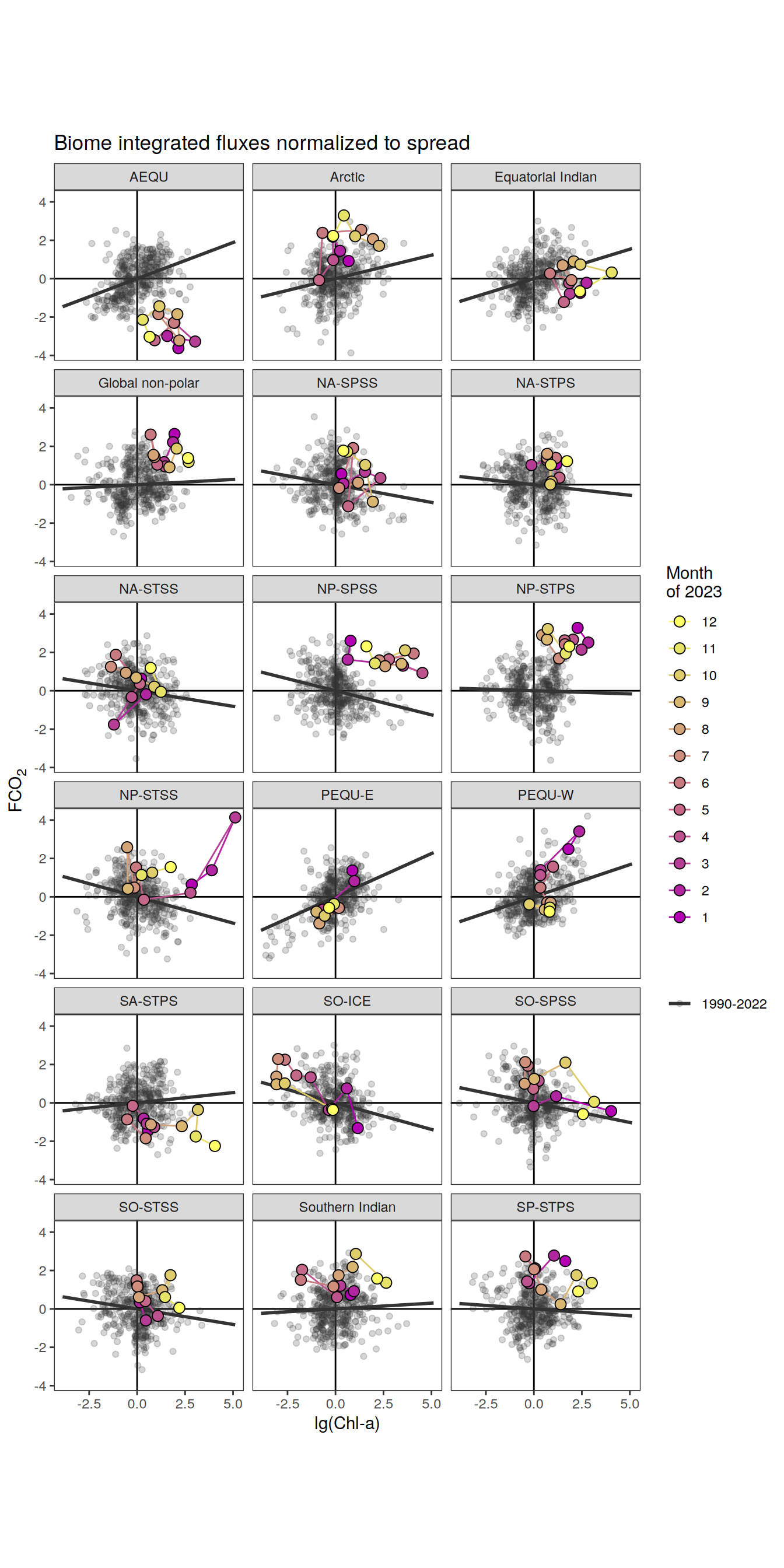
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
| a60be97 | jens-daniel-mueller | 2024-06-12 |
| 5c52cd2 | jens-daniel-mueller | 2024-06-12 |
| c69cb32 | jens-daniel-mueller | 2024-06-11 |
| 03d196b | jens-daniel-mueller | 2024-06-11 |
| a3743ec | jens-daniel-mueller | 2024-05-25 |
| 009791f | jens-daniel-mueller | 2024-05-14 |
| cd94a01 | jens-daniel-mueller | 2024-05-06 |
| 3bd3bfb | jens-daniel-mueller | 2024-04-17 |
fCO2 decomposition
biome_mask <-
bind_rows(
biome_mask,
biome_mask %>%
filter(!str_detect(biome, "SO-SPSS|SO-ICE|Arctic")) %>%
mutate(biome = "Global non-polar")
)
pco2_product_biome_monthly_fCO2_decomposition <-
full_join(pco2_product_map_monthly_fCO2_decomposition,
biome_mask,
relationship = "many-to-many") %>%
group_by(year, month, biome, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup() %>%
drop_na()
pco2_product_biome_annual_fCO2_decomposition <-
pco2_product_biome_monthly_fCO2_decomposition %>%
group_by(year, biome, name) %>%
summarise(resid = mean(resid)) %>%
ungroup()
pco2_product_biome_monthly_fCO2_decomposition %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
p_season(title = paste("Anomalies from predicted monthly mean"))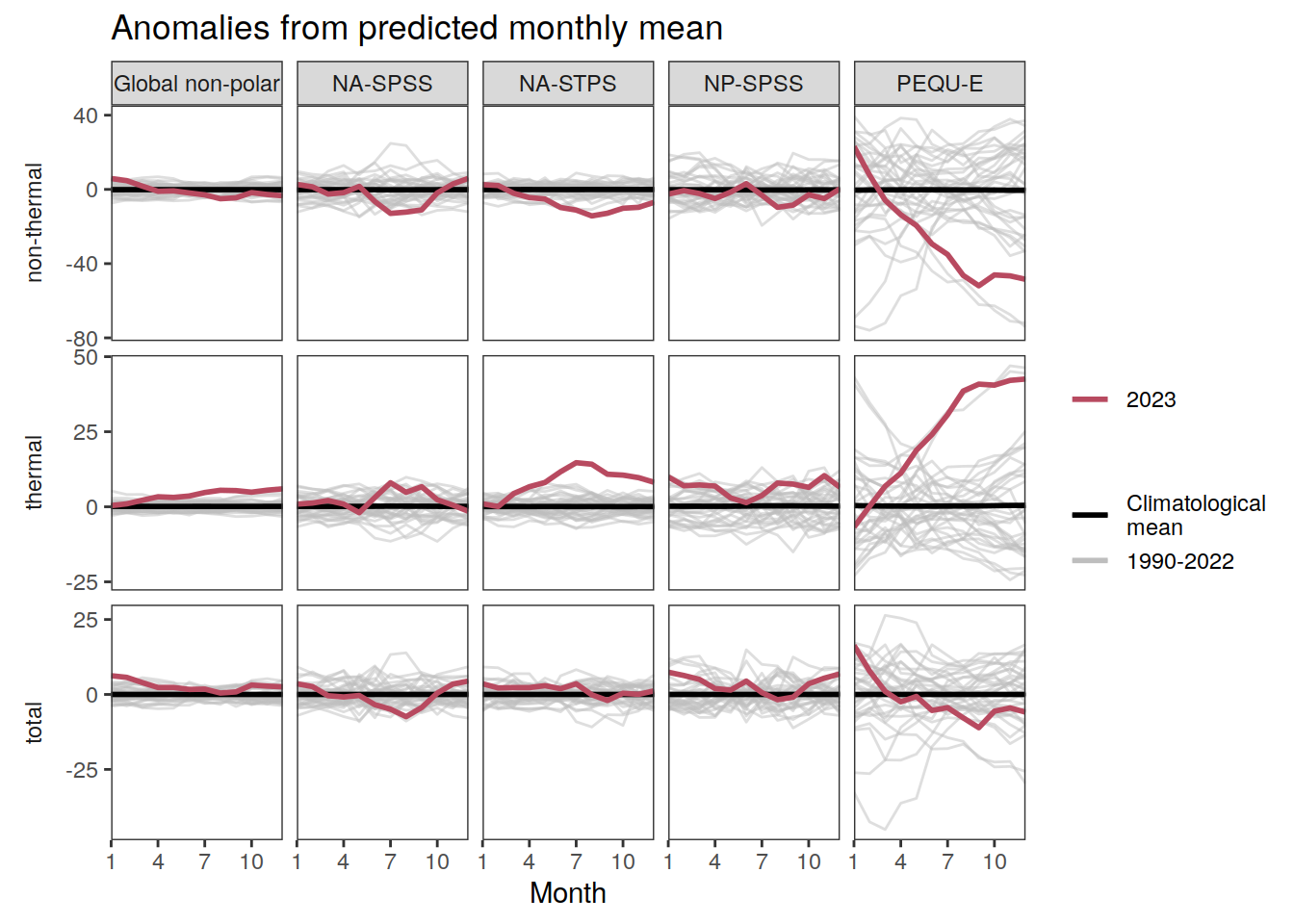
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| f6a4369 | jens-daniel-mueller | 2024-07-01 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
Flux attribution
Seasonal
pco2_product_biome_monthly_flux_attribution <-
full_join(pco2_product_map_monthly_flux_attribution,
biome_mask,
relationship = "many-to-many") %>%
group_by(year, month, biome, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup() %>%
drop_na()
pco2_product_biome_monthly_flux_attribution_total <-
full_join(pco2_product_map_monthly_anomaly %>%
filter(name == "fgco2") %>%
mutate(name = "resid_fgco2"),
biome_mask,
relationship = "many-to-many") %>%
group_by(year, month, biome, name) %>%
summarise(resid = mean(resid, na.rm = TRUE)) %>%
ungroup() %>%
drop_na()
pco2_product_biome_monthly_flux_attribution <-
bind_rows(
pco2_product_biome_monthly_flux_attribution,
pco2_product_biome_monthly_flux_attribution_total
)
pco2_product_biome_annual_flux_attribution <-
pco2_product_biome_monthly_flux_attribution %>%
group_by(year, biome, name) %>%
summarise(resid = mean(resid)) %>%
ungroup()
pco2_product_biome_monthly_flux_attribution %>%
filter(year == 2023,
biome %in% c("Global non-polar", key_biomes)) %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_path(
aes(month, resid)
) +
geom_point(
aes(month, resid),
shape = 21,
alpha = 0.5,
col = "grey30"
) +
scale_y_continuous(breaks = seq(-10,10,0.2)) +
scale_x_continuous(position = "top", breaks = seq(1,12,3)) +
labs(y = labels_breaks(unique("fgco2"))$i_legend_title) +
facet_grid(biome ~ name,
labeller = labeller(name = x_axis_labels),
scales = "free_y",
space = "free_y",
switch = "x") +
theme(
legend.title = element_blank(),
axis.title.y = element_markdown(),
strip.text.x.bottom = element_markdown(),
strip.placement = "outside",
strip.background.x = element_blank(),
legend.position = "top"
)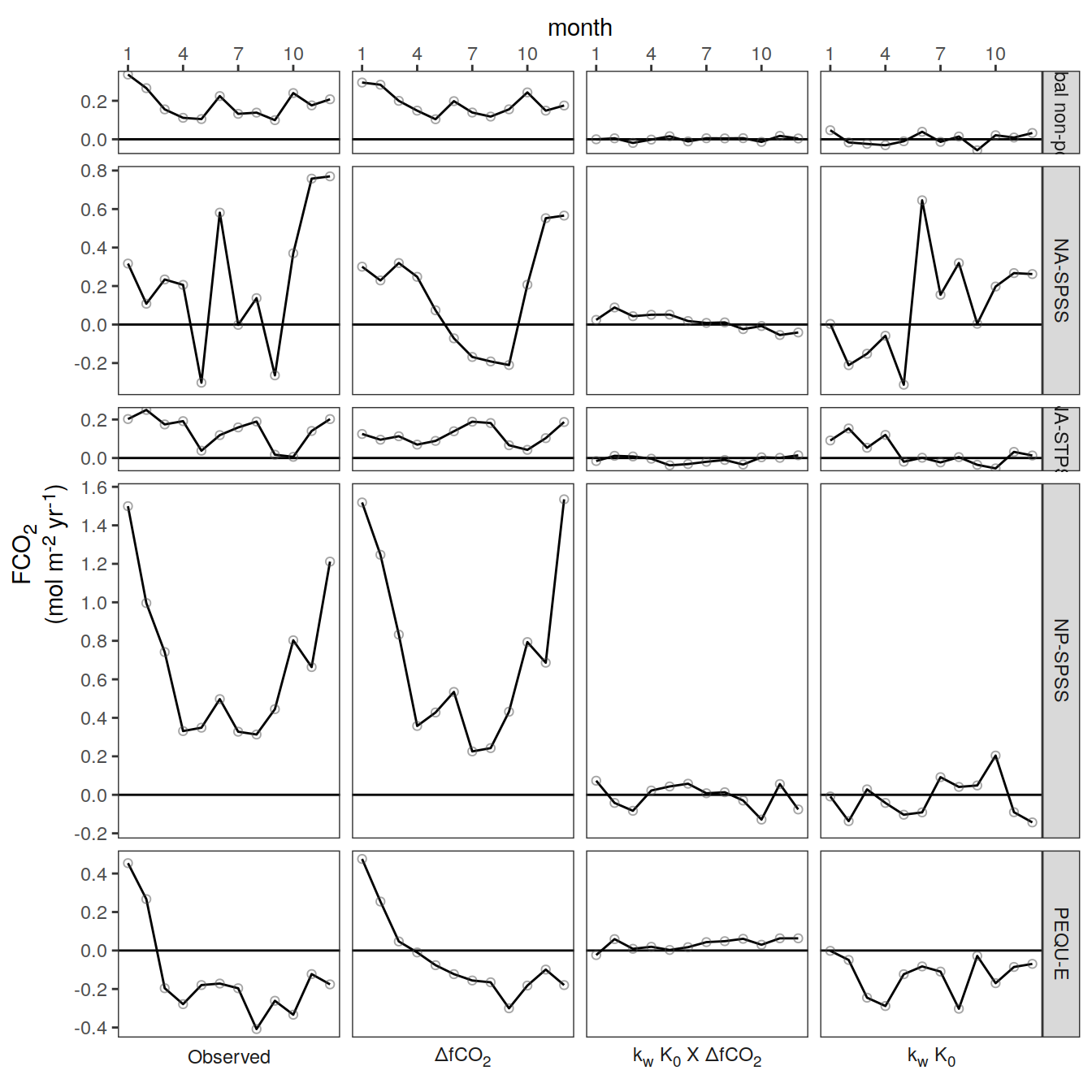
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| f6a4369 | jens-daniel-mueller | 2024-07-01 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
pco2_product_biome_monthly_flux_attribution %>%
filter(biome %in% c("Global non-polar", key_biomes)) %>%
p_season(title = paste("Anomalies from predicted monthly mean"))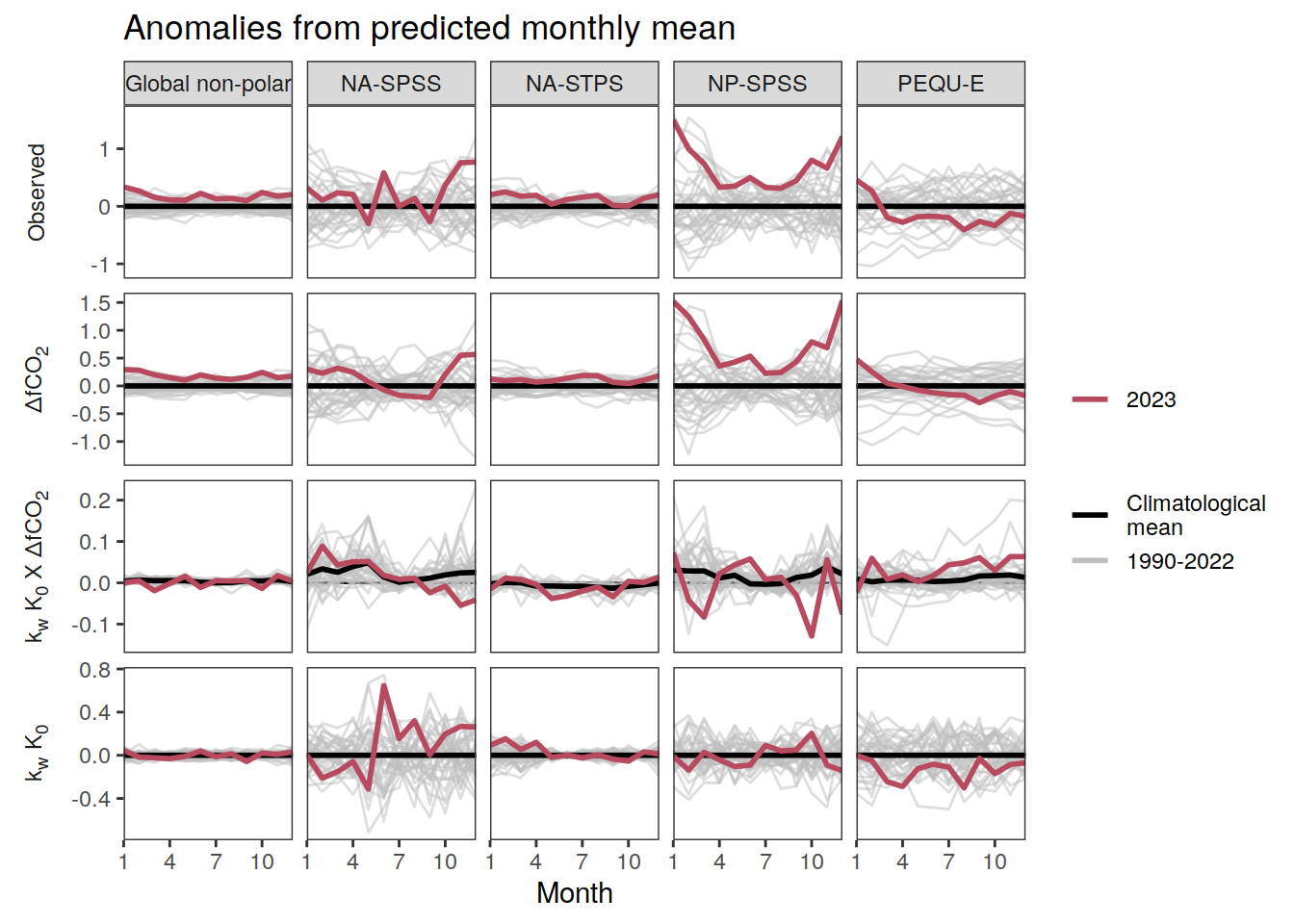
| Version | Author | Date |
|---|---|---|
| dc38c97 | jens-daniel-mueller | 2025-02-26 |
| 2ea5f0a | jens-daniel-mueller | 2025-02-25 |
| 698803c | jens-daniel-mueller | 2025-02-25 |
| f7331c3 | jens-daniel-mueller | 2025-02-23 |
| 2636a99 | jens-daniel-mueller | 2025-02-21 |
| 551f825 | jens-daniel-mueller | 2024-09-09 |
| 7c52bc2 | jens-daniel-mueller | 2024-08-23 |
| f6a4369 | jens-daniel-mueller | 2024-07-01 |
| a039cda | jens-daniel-mueller | 2024-06-26 |
Annual
pco2_product_biome_annual_flux_attribution <-
full_join(
pco2_product_biome_annual_flux_attribution %>%
filter(year == 2023) %>%
select(-year),
pco2_product_biome_annual_flux_attribution %>%
filter(year != 2023) %>%
group_by(biome, name) %>%
summarise(resid_mean = mean(abs(resid))) %>%
ungroup()
)
# pco2_product_biome_annual_flux_attribution %>%
# filter(biome %in% c("Global non-polar", key_biomes)) %>%
# mutate(product == "pco2 product") %>%
# group_split(product) %>%
# # head(1) %>%
# map(
# ~ ggplot(data = .x) +
# geom_col(aes("x", resid),
# position = "dodge2") +
# geom_col(
# aes(
# "x",
# resid_mean * sign(resid),
# col = paste0("Mean\nexcl.",2023)
# ),
# position = "dodge2",
# fill = "transparent"
# ) +
# labs(y = labels_breaks(unique("fgco2"))$i_legend_title,
# title = .x$biome) +
# facet_grid(
# biome~name,
# labeller = labeller(name = x_axis_labels),
# switch = "x"
# ) +
# scale_color_grey() +
# theme(
# legend.title = element_blank(),
# axis.text.x = element_blank(),
# axis.ticks.x = element_blank(),
# axis.title.x = element_blank(),
# axis.title.y = element_markdown(),
# strip.text.x.bottom = element_markdown(),
# strip.placement = "outside",
# strip.background.x = element_blank(),
# legend.position = "top"
# )
# )pco2_product_biome_annual_flux_attribution %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_annual_flux_attribution.csv"
)
)
pco2_product_biome_monthly_flux_attribution %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_monthly_flux_attribution.csv"
)
)
pco2_product_biome_annual_fCO2_decomposition %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_annual_fCO2_decomposition.csv"
)
)
pco2_product_biome_monthly_fCO2_decomposition %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_biome_monthly_fCO2_decomposition.csv"
)
)
rm(
pco2_product_biome_annual_flux_attribution,
pco2_product_biome_monthly_flux_attribution,
pco2_product_biome_annual_fCO2_decomposition,
pco2_product_biome_monthly_fCO2_decomposition
)
gc()Zonal mean sections
The following analysis is available for GOBMs only.
Annual means
2023 anomaly
pco2_product_zonal_mean_annual <- pco2_product_zonal_mean %>%
pivot_longer(-c(region, depth, lat, time, year, month)) %>%
group_by(region, lat, depth, year, name) %>%
summarise(value = mean(value)) %>%
ungroup() %>%
drop_na() %>%
mutate(region = str_to_title(region))
pco2_product_zonal_mean_annual_anomaly <-
pco2_product_zonal_mean_annual %>%
anomaly_determination(region, lat, depth)
pco2_product_zonal_mean_annual_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(3) %>%
map(
~ ggplot(data = .x) +
geom_contour_filled(aes(lat, depth, z = resid)) +
scale_fill_discrete_divergingx(name = labels_breaks(.x %>% distinct(name))$i_legend_title) +
guides(fill = guide_colorsteps(
barheight = unit(8, "cm"),
show.limits = TRUE
)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50,100,200,400)) +
scale_x_continuous(breaks = seq(-100, 100, 20)) +
coord_cartesian(expand = 0) +
facet_wrap( ~ region, ncol = 1) +
labs(y = "Depth (m)") +
theme(legend.title = element_markdown())
)pco2_product_zonal_mean_annual_anomaly %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_zonal_mean_sections_annual.csv"
)
)Biome profiles
The following analysis is available for GOBMs only.
Annual means
2023 anomaly
pco2_product_profiles_annual <- pco2_product_profiles %>%
pivot_longer(-c(biome, depth, time, year, month)) %>%
group_by(biome, depth, year, name) %>%
summarise(value = mean(value)) %>%
ungroup() %>%
drop_na()
pco2_product_profiles_annual_anomaly <-
pco2_product_profiles_annual %>%
anomaly_determination(biome, depth)
pco2_product_profiles_annual_anomaly %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_path(aes(resid, depth, group = year), col = "grey30", alpha = 0.5) +
geom_path(data = .x %>% filter(year == 2023),
aes(resid, depth, col = as.factor(year)),
linewidth = 1) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50,100,200,400)) +
facet_wrap( ~ biome) +
labs(y = "Depth (m)",
x = labels_breaks(.x %>% distinct(name))$i_legend_title) +
theme(legend.title = element_blank(),
axis.title.x = element_markdown())
)pco2_product_profiles_annual_anomaly %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_profiles_annual.csv"
)
)Monthly means
2023 anomaly
pco2_product_profiles_monthly <- pco2_product_profiles %>%
pivot_longer(-c(biome, depth, time, year, month)) %>%
group_by(biome, depth, year, month, name) %>%
summarise(value = mean(value)) %>%
ungroup() %>%
drop_na()
pco2_product_profiles_monthly_anomaly <-
pco2_product_profiles_monthly %>%
anomaly_determination(biome, depth, month)
pco2_product_profiles_monthly_anomaly %>%
filter(year == 2023) %>%
group_split(name) %>%
# head(1) %>%
map(
~ ggplot(data = .x) +
geom_vline(xintercept = 0) +
geom_path(aes(resid, depth, col = as.factor(month)),
linewidth = 1) +
scale_color_scico_d(palette = "hawaii") +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(50, 100, 200, 400)) +
facet_wrap(~ biome,
scales = "free_x") +
labs(y = "Depth (m)",
x = labels_breaks(.x %>% distinct(name))$i_legend_title) +
theme(legend.title = element_blank(),
axis.title.x = element_markdown())
)pco2_product_profiles_monthly_anomaly %>%
write_csv(
paste0(
"../data/",
"fCO2-Residual",
"_",
"2023",
"_profiles_monthly.csv"
)
)
sessionInfo()R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: openSUSE Leap 15.6
Matrix products: default
BLAS/LAPACK: /usr/local/OpenBLAS-0.3.28/lib/libopenblas_haswellp-r0.3.28.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Zurich
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggh4x_0.3.0 scales_1.3.0 cmocean_0.3-2
[4] ggtext_0.1.2 broom_1.0.7 khroma_1.14.0
[7] ggnewscale_0.5.0 stars_0.6-7 abind_1.4-8
[10] terra_1.8-5 sf_1.0-19 rnaturalearth_1.0.1
[13] geomtextpath_0.1.4 colorspace_2.1-1 marelac_2.1.11
[16] shape_1.4.6.1 ggforce_0.4.2 metR_0.16.0
[19] scico_1.5.0 patchwork_1.3.0 collapse_2.0.18
[22] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
[25] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5
[28] tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.1
[31] tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] DBI_1.2.3 rlang_1.1.4 magrittr_2.0.3
[4] git2r_0.35.0 e1071_1.7-16 compiler_4.4.2
[7] mgcv_1.9-1 getPass_0.2-4 systemfonts_1.1.0
[10] callr_3.7.6 vctrs_0.6.5 pkgconfig_2.0.3
[13] crayon_1.5.3 fastmap_1.2.0 backports_1.5.0
[16] ncmeta_0.4.0 labeling_0.4.3 utf8_1.2.4
[19] promises_1.3.2 rmarkdown_2.29 markdown_1.13
[22] tzdb_0.4.0 ps_1.8.1 oce_1.8-3
[25] gsw_1.2-0 bit_4.5.0 xfun_0.49
[28] cachem_1.1.0 jsonlite_1.8.9 later_1.4.1
[31] tweenr_2.0.3 parallel_4.4.2 R6_2.5.1
[34] RColorBrewer_1.1-3 bslib_0.8.0 stringi_1.8.4
[37] jquerylib_0.1.4 Rcpp_1.0.13-1 knitr_1.49
[40] seacarb_3.3.3 Matrix_1.7-1 splines_4.4.2
[43] httpuv_1.6.15 timechange_0.3.0 tidyselect_1.2.1
[46] RNetCDF_2.9-2 rstudioapi_0.17.1 yaml_2.3.10
[49] codetools_0.2-20 processx_3.8.4 lattice_0.22-6
[52] withr_3.0.2 evaluate_1.0.1 rnaturalearthdata_1.0.0
[55] units_0.8-5 proxy_0.4-27 polyclip_1.10-7
[58] xml2_1.3.6 pillar_1.9.0 whisker_0.4.1
[61] CFtime_1.4.1 KernSmooth_2.23-24 checkmate_2.3.2
[64] generics_0.1.3 vroom_1.6.5 rprojroot_2.0.4
[67] hms_1.1.3 commonmark_1.9.2 munsell_0.5.1
[70] class_7.3-22 glue_1.8.0 tools_4.4.2
[73] data.table_1.16.2 fs_1.6.5 grid_4.4.2
[76] nlme_3.1-166 cli_3.6.3 SolveSAPHE_2.1.0
[79] textshaping_0.4.0 fansi_1.0.6 viridisLite_0.4.2
[82] gtable_0.3.6 sass_0.4.9 digest_0.6.37
[85] classInt_0.4-10 farver_2.1.2 memoise_2.0.1
[88] htmltools_0.5.8.1 lifecycle_1.0.4 httr_1.4.7
[91] here_1.0.1 gridtext_0.1.5 bit64_4.5.2
[94] MASS_7.3-61